Key Points
HY, historically considered an initiating event in MM, can be preceded by deletions involving driver genes.
Deletions within large chromosomal gains represent a novel mechanism that impacts both driver gene expression and clinical outcomes.
Visual Abstract
Acquisition of a hyperdiploid (HY) karyotype or immunoglobulin heavy chain (IgH) translocations are considered key initiating events in multiple myeloma (MM). To explore if other genomic events can precede these events, we analyzed whole-genome sequencing data from 1173 MM samples. By integrating molecular time and structural variants within early chromosomal duplications, we indeed identified pregain deletions in 9.4% of patients with an HY karyotype without IgH translocations, challenging acquisition of an HY karyotype as the earliest somatic event. Remarkably, these deletions affected tumor suppressor genes (TSGs) and/or oncogenes in 2.4% of patients with an HY karyotype without IgH translocations, supporting their role in MM pathogenesis. Furthermore, our study points to postgain deletions as novel driver mechanisms in MM. Using multiomics approaches to investigate their biologic impact, we found associations with poor clinical outcome in newly diagnosed patients and profound effects on both the oncogene and TSG activity despite the diploid gene status. Overall, this study provides novel insights into the temporal dynamics of genomic alterations in MM.
Introduction
Multiple myeloma (MM) is the second most common hematologic cancer and is characterized by aberrant proliferation of plasma cells in the bone marrow (BM).1,2 MM is always preceded by precursor conditions, including monoclonal gammopathy of undetermined significance and smoldering MM.2-6 The life history and progression of MM through these clinically recognized precursor conditions is believed to start as a consequence of a complex and diverse genomic evolution in the germinal center (GC), initiated by acquisition of a hyperdiploid karyotype (HY) (ie, multiple trisomies on odd-numbered chromosomes) or translocations between the immunoglobulin heavy chain locus (IgH) and distinct oncogenes, such as CCND1.1,7 This understanding was solidified more than 2 decades ago through the integration of fluorescence in situ hybridization and gene expression analysis.8-15 Furthermore, according to Maura et al,16 HY in patients with MM who lack canonical IgH translocations may manifest up to 30 to 40 years before diagnosis. However, the potential acquisition of other somatic events before HY and canonical IgH translocations has neither been considered nor explored. Although nonsynonymous mutations in driver genes that precede large chromosomal gains in patients with HY MM have been documented,17 the resolution of whole exome sequencing in that study was inadequate for a comprehensive characterization of the early nature of these large chromosomal gains.
The recombination-activating genes (RAG) complex is known to play a key role in the variable diversity joining (VDJ) recombination during the early phases of B-cell maturation and distinct B-cell tumors.18,19 Machado et al recently used whole-genome sequencing (WGS) data from normal B cells to reveal sporadic RAG-mediated deletions and structural variants (SV) that span the genome of both normal B-naive and memory cells, including loci of tumor suppressor genes (TSGs).20 These findings are highly suggestive of genomic events that can be acquired in normal B cells before any neoplastic initiation. Interestingly, recent studies have suggested that the RAG complex might be involved in a fraction of MM canonical IgH translocations in BM during VDJ recombination, challenging the conventional belief that MM always originates in the GC.21,22 The RAG complex is known to be responsible for 2 key cancer initiating events in other B-cell tumors, namely the t(11;14)(CCND1;IGH) translocation in mantle cell lymphoma (MCL) and the t(14;18)(IGH;BCL2) translocation in diffuse large B-cell lymphoma (DLBCL) and follicular lymphoma (FL).18-20,23,24 The potential role of the RAG complex in the etiology of these lymphomas has been established based on 2 characteristic features. First, because both IgH translocation breakpoints are localized within the VDJ region, it fits with the involvement of RAG in the translocation event. Second, the presence of recombination signal sequence (RSS) motifs near the translocation breakpoints further supports the involvement of the RAG complex in the translocation process.19 In MM, ∼20% of t(11;14)(CCND1;IGH) breakpoints are localized within the VDJ region, suggesting that aberrant RAG complex activity may be responsible for their acquisition, similar to what is observed in MCL.22,24 However, because of the lack of thorough investigations of RSS motifs, the pathogenetic role of RAG in MM remains a subject of debate.
To explore the presence, prevalence, and biologic significance of genomic events that occur before the acquisition of trisomies in HY patients without canonical IgH translocations (HY/IgH-neg), we developed an innovative computational workflow that allowed us to time deletions within large chromosomal gains. Our investigations unveiled that in 2.4% of HY/IgH-neg patients with newly diagnosed MM (NDMM), a deletion that causes dysregulation in TSGs or oncogenes can precede the occurrence of HY trisomies, which are considered initiating events. Of note, our study also provides new findings of monoallelic deletions that are acquired after large chromosomal gains (ie, postgain deletions) and that have clinical prognostic value. Integrating bulk RNA sequencing (RNAseq) and single-cell assay for transposase-accessible chromatin using sequencing (scATACseq), we show that these deletions, which revert the copy number from trisomic to diploid, have a profound biologic influence on the expression of both oncogenes and TSGs.
Methods
Patients
This study, which was conducted in adherence with the Declaration of Helsinki, received approval from the Memorial Sloan Kettering Cancer Center (MSKCC) and the Heidelberg University. WGS from 421 samples were generated at MSKCC (n = 60) and Heidelberg University (n = 361). Details about the sorting, WGS, RNA, scATACseq, and analytical approaches can be found in the supplemental Methods, available on the Blood website. Patients who harbored at least 2 large chromosomal gains on odd-numbered chromosomes (3, 5, 7, 9, 11, 15, 19, or 21) were defined as HY.25
Molecular time analysis
To time the acquisition of large chromosomal gains, we used the R package mol_time (https://github.com/UM-Myeloma-Genomics/mol_time).26 This approach enabled the estimation of the relative timing of large chromosomal gains (>1 Mb), clonal chromosomal gains (3;1 or 4:1), trisomies (3:1), copy neutral loss of heterozygosity (2:0), and amplification (4:1). The estimation relied on the purity-corrected ratio between duplicated clonal mutations. Clonal single base substitutions (SBSs) were categorized into duplicated or nonduplicated based on the variant allele frequency (VAF) corrected for cancer purity. The latter was estimated by combining the purity estimates from both the ASCAT27 and the SBS VAF density and distribution within clonal diploid regions. Segments with >50 clonal mutations, excluding immunoglobulin loci and kataegis events, were considered for copy number variant (CNV) analysis. Tetrasomies (2:2) were excluded because of the challenge of determining whether both chromosomal gains occurred in close temporal succession. The relative molecular time of each gained copy number abnormality segment was estimated using the described approach26 with distinction between chromosomal gains that occurred in the same time window and those that occurred in different time windows.
SV-mediated pregain and postgain deletion analysis
Pregain analysis consisted of identifying clonal CNVs supported by SVs that caused a copy number drop from 3:1 (total alleles:minor allele) to 1:0 on large clonal early gains, determined by molecular time. Only duplications in which ≥40% of a chromosome arm was gained were included. Pregain deletion analysis only considered early gains as determined by molecular time for NDMM in the MSKCC and German cohorts. For the Multiple Myeloma Research Foundation (MMRF) NDMM samples, we considered all HY chromosomes and assumed that the gains were acquired at the same time. This decision was based on the data reported by Maura et al16,26 that showed that 90% of HY have large trisomy on odd chromosomes that are acquired within the same time window. In contrast, for the postgain deletion analysis, we considered all NDMM and relapsed refractory MM (RRMM) samples with a CNV supported by SVs that caused a copy number drop from 3:1 to 2:1 on large clonal gains. Only duplications in which ≥40% of a chromosome arm was gained were included. For both pre- and postgain deletions, CNVs and SVs were linked if at least 1 SV breakpoint was within 50 kb of the CNV breakpoint.28 SV-mediated pregain deletions involving IGH, IGL, or IGK regions were removed from our analysis.
Driver analysis
We determined whether pregain and postgain deletions affected the transcriptional levels of oncogenes and TSGs. This catalog of driver genes was generated by integrating the Catalogue of Somatic Mutations in Cancer (COSMIC) census with previously defined MM drivers (supplemental Table 1).26,29 Genes were defined as either an oncogene or a TSG based on COSMIC definitions. Any oncogene or TSG with a double annotation according to COSMIC was deemed 1 or the other in the context of the hematologic malignancies, including MM, MCL, DLBCL, FL, and leukemias. As summarized in supplemental Figure 1, loss-of-function (LOF) events were defined as reduced gene expression levels (ie, within in the lower quartile of expression when compared with patients in the respective cohort) of any TSG within 50 kb upstream or downstream of the deleted segment. Gain-of-function of an oncogene was defined as any oncogene defined outside of a deletion footprint of a pre- or postgain deletion up to 500 kb on either end of the CNV deletion segment breakpoint that led to overexpression of genes (ie, the upper quartile of expression when compared with patients in the respective cohort). Fusion genes created by pre- or postgain deletions were defined as a driver if as at least 1 TSG/oncogene was joined to another gene. A distance of 2 kb on either end of a CNV deletion breakpoint was allowed. Only fusions with expression greater than the median expression of other patients in the same cohort were considered. These criteria were employed to maintain consistency with our previous research, to take into account the average size of topologically associated domain (500 kb), and to correct possible discrepancy between copy number and SV data.28,30
Survival analysis
Survival analysis was performed using the R package Survival (https://cran.r-project.org/web/packages/survival/index.html). The pregain Kaplan-Meier (KM) analysis compared samples with at least 1 pregain deletion event with those without a pregain deletion event. The postgain KM analysis compared samples with at least 3 postgain events with those with 2 events or less. A postgain deletion analysis was also conducted as a linear feature using the coxph R package. Both pregain and postgain KM analyses only considered NDMM samples with progression-free survival (PFS) or overall survival (OS) clinical information available with separate analyses conducted for all samples and HY/IgH-neg samples. KM analyses only considered the German and MSKCC cohorts. Coxph linear analysis of postgain samples were conducted with consideration of the number of postgain events, International Staging System (ISS) status (I/II or III), and the presence or absence of at least 1 of the following cytogenetic abnormalities: TP53 del or mutation, t(4;14)[NSD2], t(14;16)[MAF], t(14;20)[MAFB].
RAG motif analysis
For RAG analysis, we investigated all nucleotide sequence ±50 positions around each SV breakpoint on either end. The sequences were run through the RSS motif tool (https://www.itb.cnr.it/rss/analyze.html). RAG+ motifs were defined as having 1 positive RSS23 or RSS12 on either end of the SV breakpoint.20,23
Statistical analysis and plotting
Proportional testing was performed using the Wilcoxon and Fisher exact tests to compare the median of a continuous variable or the distribution of discrete variables across groups when appropriate. All P values are 2-sided if not specified otherwise. The KM estimator was used to calculate time-to-event distributions. PFS was measured from the date of start of treatment to the date of progression or death, whichever occurred first. All plots and figures were generated using R-Studio Version 2023.09.1+494 (2023.09.1+494), Adobe Illustrator, and Biorender.
Results
Defining deletion events acquired before early large chromosomal gains
To address the hypothesis that MM cells acquire genomic driver aberrations before established initiating events, we developed a chronologic analytical model based on the integration of single nucleotide variants (SNV), SV, and CNV (Figure 1). First, we used molecular time, calculated as the corrected ratio between duplicated and nonduplicated clonal mutations within large chromosomal gains (ie, ≥40% of the chromosome arm), to pinpoint the earliest clonal set of chromosomal gains in each patient.26,31,32 Next, we examined SV-mediated deletions within the earliest set of gains in each patient. Each of these deletions was categorized into 1 of 3 classes, namely (1) the deletion occurred on the allele that will later be duplicated, which led to a CNV jump from 3:1 to 1:0 (total alleles:minor alleles), called a pregain deletion (Figure 1A); (2) loss of 1 of the duplicated alleles after the gain, causing a CNV shift from 3:1 to 2:1, called a postgain deletion (Figure 1B); and (3) the deletion occurred on the minor, nonduplicated allele, causing a CNV jump from 3:1 to 2:0 and leading to a loss of heterozygosity. This last scenario could be explained by both pre- and postgain events and was therefore excluded from our analytical workflow (Figure 1C). The analysis of pregain deletions was centered around the earliest set of gains in HY/IgH-neg patients with NDMM, which was believed to be the origin of the disease in this subgroup (Figure 1D-F).16,26,32 In contrast, for the analysis of postgain deletions, we considered all patients, including patients with RRMM (Figure 1G). Finally, we integrated WGS and RNAseq data (n = 830) to predict the biologic impact of pre- and postgain deletions.
Workflow for detecting pregain and postgain deletions in patients with MM. Cartoons summarizing a (A) pregain deletion event, (B) postgain deletion event, and (C) pregain deletion and postgain deletion events involving a loss-of-heterozygosity. Numbers (ie, 3:1) indicate the number of total alleles to the number of minor alleles. (D) Example of a copy-number plot of a HY/IgH-neg patient with newly diagnosed MM. (E) Molecular time output for the patient’s large chromosomal gains. (F) Copy number plot zoomed in on pregain deletion involving the TSG TCF3. (G) Example of postgain deletion near the oncogene PIK3CA. In panels F-G, the horizontal black and orange lines represent the total copy number and the minor allele, respectively. The vertical line represents SV deletions. (H) Workflow for identifying early pregain (yellow) and postgain (green) deletion events mediated by SV in study cohorts.
Workflow for detecting pregain and postgain deletions in patients with MM. Cartoons summarizing a (A) pregain deletion event, (B) postgain deletion event, and (C) pregain deletion and postgain deletion events involving a loss-of-heterozygosity. Numbers (ie, 3:1) indicate the number of total alleles to the number of minor alleles. (D) Example of a copy-number plot of a HY/IgH-neg patient with newly diagnosed MM. (E) Molecular time output for the patient’s large chromosomal gains. (F) Copy number plot zoomed in on pregain deletion involving the TSG TCF3. (G) Example of postgain deletion near the oncogene PIK3CA. In panels F-G, the horizontal black and orange lines represent the total copy number and the minor allele, respectively. The vertical line represents SV deletions. (H) Workflow for identifying early pregain (yellow) and postgain (green) deletion events mediated by SV in study cohorts.
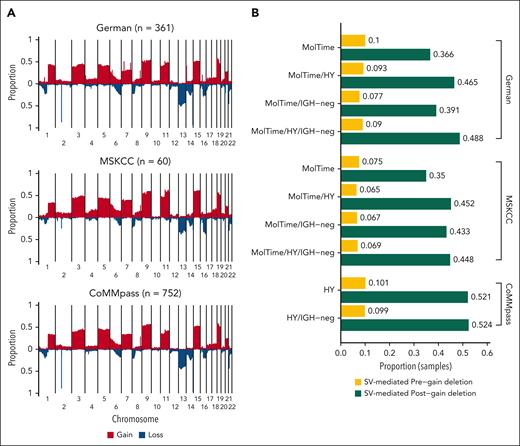
We applied this workflow to 3 independent cohorts of patients with NDMM with WGS data (Figures 1H and 2A; supplemental Tables 2-3). For 2 cohorts, high-coverage WGS data (85.7×; range, 63.1× to 176.6×) were available. The first cohort (here called German) included 263 transplant-eligible patients with NDMM who were treated in Germany, 256 of whom were enrolled in the GMMG-HD6 trial in which patients were treated with VRd (bortezomib, lenalidomide dexamethasone, n = 68) or Elo-VRd (elotuzumab, bortezomib, lenalidomide dexamethasone, n = 187).25,33 Second, the MSKCC cohort (n = 60) included patients who were treated with KRd (carfilzomib, lenalidomide, and dexamethasone) (n = 16) or Daratumumab-KRD (n = 44) within 2 phase 2 clinical trials.34-36 For validation, we used the long-insert, low-coverage (4× to 8×) WGS MMRF CoMMpass study of NDMM (n = 752) samples.28 Given that the MMRF CoMMpass WGS data were long-insert and low-coverage data, we were unable to reliably generate molecular time estimates to determine the earliest gains. Therefore, for the MMRF CoMMpass cohort, we restricted our pregain deletion analysis only to HY/IgH-neg samples and assumed all large gains on HY odd-numbered chromosomes were acquired in the same time-window (3, 5, 7, 9, 11, 15, 19, 21) (Figure 1D). An additional 98 WGS data from patients with RRMM treated in Germany were included to investigate the impact of postgain deletions.37,38
Prevalence and biologic impact of deletion acquired before large chromosomal gains
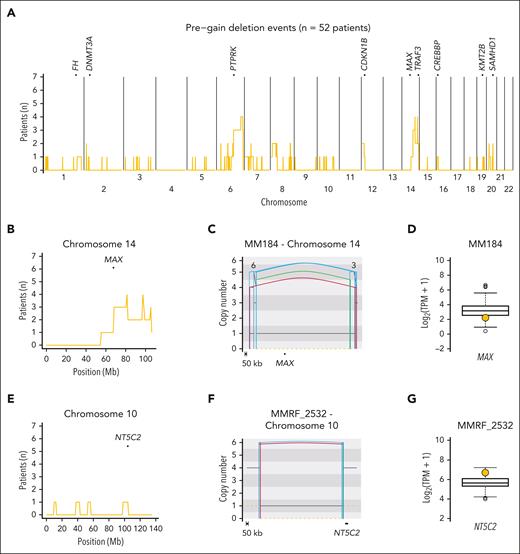
Molecular time data were successfully generated for 249 of 323 (77%) patients with NDMM using high-coverage WGS. Only 10.9% of HY samples were not included in the molecular time analysis because of low mutational burden within duplicated regions (supplemental Methods). Restricting our analysis to patients with HY/IgH-neg, SV-mediated deletions acquired before early large clonal chromosomal gains were identified in 6.9% and 9.8% of patients with NDMM in the MSKCC and German cohorts, respectively (Figure 2B; supplemental Table 4). In the low-coverage validation group, 9.9% HY/IgH-neg patients in the CoMMpass cohort showed pregain deletions (Figure 2B). In total, we detected 89 SV–mediated pregain deletion events among 52 of 552 (9.4%) HY/IgH-neg patients across the 3 NDMM series (supplemental Table 5). The median size of pregain deletions was 25 522 636 bp (range, 1676-78 234 366 bp). To investigate the impact of these events on MM driver genes (supplemental Methods; supplemental Figure 1), we restricted our analysis to 39 of 52 (75%) of these patients with matched WGS/RNAseq data (69/89, 77.5% pregain SV events). Among these events, 14 of 69 (20.3%) pregain deletions were responsible for the downregulation of at least 1 TSG in 13 of 39 (33%) patients, including known MM drivers such as MAX, TRAF3, and DNMT3A (Figure 3A-D). In contrast 3 of 69 (4.35%) events in 2 of 39 (5.1%) patients were associated with upregulation of at least 1 oncogene, such as PLCG1, NT5C2, and SRC. No fusion gene was detected (Figure 3E-G). Overall, 2.4% of HY/IgH-neg patients with available WGS and RNAseq data had pregain deletions that affected TSGs or oncogenes.
Cohort-wide copy-number profile and pre-/postgain deletions in MM. (A) Cumulative copy number plot summarizing the copy number distribution across the 3 series of MM. (B) Proportion of samples with SV-mediated pregain (yellow) and postgain (green) deletions according to different categories. MolTime: Samples with early gains determined by molecular time analysis. Pregain deletions include only NDMM samples, whereas postgain deletions include NDMM and RRMM samples.
Cohort-wide copy-number profile and pre-/postgain deletions in MM. (A) Cumulative copy number plot summarizing the copy number distribution across the 3 series of MM. (B) Proportion of samples with SV-mediated pregain (yellow) and postgain (green) deletions according to different categories. MolTime: Samples with early gains determined by molecular time analysis. Pregain deletions include only NDMM samples, whereas postgain deletions include NDMM and RRMM samples.
Prevalence and distribution of pregain deletions in HY patients with newly diagnosed myeloma. (A) Cumulative distribution plot of pregain deletions in 55 HY/IgH-neg patients with NDMM. Top of plot shows TSGs that were enriched more than the median number of patients per 1 Mb bin with a pregain deletion on the given chromosome. (B) Enlarged distribution of pregain deletions on chromosome 14 involving the TSG MAX. (C) Copy number plot showing pregain deletion, including MAX, in patient MM184. (D) RNAseq transcript levels of patients with NDMM from the German cohort with patient MM184 highlighted in yellow who showed a clear MAX downregulation. (E) Enlarged distribution of pregain deletions per 1 Mb bin on chromosome 10 around the oncogene NT5C2. (F) Copy number plot showing a pregain deletion near NT5C2 in patient MMRF_2532. (G) RNAseq transcript levels of patients with NDMM from MMRF with patient MMRF_2532 highlighted in yellow, showing a clear overexpression of NT5C2. In panels C, F, the black and orange horizontal lines represent the total and minor copy number, respectively. The black, red, green, and blue vertical lines represent translocations, deletions, duplications, and inversions. For translocations, the partner chromosome is reported on the top of the line.
Prevalence and distribution of pregain deletions in HY patients with newly diagnosed myeloma. (A) Cumulative distribution plot of pregain deletions in 55 HY/IgH-neg patients with NDMM. Top of plot shows TSGs that were enriched more than the median number of patients per 1 Mb bin with a pregain deletion on the given chromosome. (B) Enlarged distribution of pregain deletions on chromosome 14 involving the TSG MAX. (C) Copy number plot showing pregain deletion, including MAX, in patient MM184. (D) RNAseq transcript levels of patients with NDMM from the German cohort with patient MM184 highlighted in yellow who showed a clear MAX downregulation. (E) Enlarged distribution of pregain deletions per 1 Mb bin on chromosome 10 around the oncogene NT5C2. (F) Copy number plot showing a pregain deletion near NT5C2 in patient MMRF_2532. (G) RNAseq transcript levels of patients with NDMM from MMRF with patient MMRF_2532 highlighted in yellow, showing a clear overexpression of NT5C2. In panels C, F, the black and orange horizontal lines represent the total and minor copy number, respectively. The black, red, green, and blue vertical lines represent translocations, deletions, duplications, and inversions. For translocations, the partner chromosome is reported on the top of the line.
A total of 31 canonical IgH translocations were found to be directly or indirectly associated with the acquisition of large chromosomal gains. When the translocations were directly linked to the gain, the molecular time of the gain was early, consistent with their early initiating role. Conversely, a late molecular time was observed when the gains were acquired after and independent from the IgH translocation. As described by Maura et al,26 canonical IgH translocations were consistently identified as the earliest event in MM pathogenesis, even when HY and IgH translocations co-occurred in the same patient. Among all these patients for whom timing of these 2 events could be determined, the deletions on the gains were always acquired after IgH translocations (supplemental Figure 2). Notably, we did not find any acquired events before the canonical IgH translocations.
In further support of the occurrence of driver events preceding early gains in HY/IgH-neg cases, we identified 5 clonal nonsynonymous mutations in MM driver genes25 that were acquired before the earliest chromosomal duplication (ie, duplicated with VAF of ∼66% once corrected for purity) (supplemental Figure 3). When compared with our previous work on the CoMMpass data set,17 the molecular time data enabled us to pinpoint the earliest set of HY gains, illustrating that nonsynonymous mutations in driver genes can indeed occur before early gains in HY/IgH-neg patients.
Finally, the presence of pregain deletions did not have a significant impact on clinical outcomes, whether considering all patients or specifically focusing on HY/IgH-neg cases (supplemental Figure 4). Overall, our data suggest that trisomies in HY patients without canonical IgH translocations can be preceded by genomic events, such as deletions, that affect the expression of TSGs or oncogenes. This is the first evidence of genomic events that can be acquired before canonical initiating events, such as HY, in MM pathogenesis.
RAG and MM pathogenesis
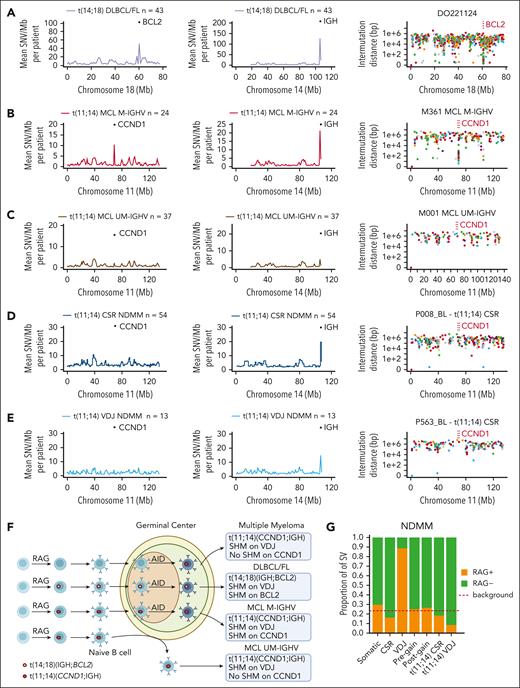
Leveraging our large series of WGS data, we aimed to define the role of the RAG complex in the acquisition of the earliest MM genomic events (ie, canonical IgH translocations and pregain deletion in HY/IgH-neg cases). Therefore, we developed an analytical workflow based on a cross-comparison between MM and other B-cell lymphomas. WGS data from patients with immunoglobulin heavy-chain variable region gene (IGHV)-unmutated MCL (n = 37) and IGHV-mutated MCL (n = 24) were included as positive controls for RAG-mediated t(11;14)(CCND1;IGH) in pre- and post-GC lymphomas, respectively. Samples with DLBCL/FL that harbored t(14;18)(IGH;BCL2) (n = 43) were included as positive controls for post-GC lymphomas that carry a RAG-mediated IgH translocation that does not involve the CCND1 gene locus.24,39 Our workflow was based on the following model: a VDJ translocation caused by RAG that was acquired before the GC-exposure to somatic hypermutation (SHM) mediated by activation-induced cytidine deaminase (AID).24 Accordingly, when a B-cell harboring a RAG-mediated VDJ translocation enters the GC, the translocated oncogene, now joined to the VDJ, will be involved in SHM with increased clustered AID-mediated mutational burden on the respective regions involved in the translocation. In line with this model, both BCL2 in DLBCL/FL and CCND1 in IGHV-mutated MCL showed clear evidence of SBS84 signature-mediated kataegis (ie, AID), a hallmark of SHM (Figure 4A-B; supplemental Figure 5A-B).40-42 In contrast, CCND1 involving the VDJ region in t(11;14)(CCND1;IGH) in both MM and IGHV-unmutated MCL did not show any evidence of SHM around CCND1 except in one patient with MM (Figure 4C-E; supplemental Figure 5C-E). Overall, this suggests that aberrant RAG activity rarely causes MM IgH translocations before the GC (Figure 4F). To further expand these findings, we examined evidence of RAG activity enrichment (ie, RSS score) across 7 different groups of SVs, namely VDJ deletions (positive control for RAG), class-switch recombination (CSR) deletions (negative control), pregain deletions, postgain deletions, those involved in t(11;14)(CCND1;IGH) with breakpoints within the CSR and VDJ regions, and all other (somatic) deletions that were not included in the previous groups (Figure 4G).19,20,43 As expected, SVs that occurred in VDJ regions of MM cells showed an enrichment for RAG activity, whereas SVs in the CSR region did not. Both pre- and postgain SV-mediated deletions did not show an enrichment for the RAG motif and neither did SVs involved with t(11;14)(CCND1;IGH). The only patient with CCND1 AID-mediated kataegis had the RAG motifs near the CCND1 breakpoints. Overall, this analysis demonstrated that RAG is rarely involved in MM initiation, neither in pregain deletions nor in the acquisition of the t(11;14)(CCND1;IGH).
Defining the germinal center origin of t(11;14)(CCND1;IGH) in MM. (A) Showing the mean SNV burden per megabase (Mb) on chromosomes 18 and 14 in DLBCL and FL samples with t(14;18)(IGH;BCL2). The rightmost plots in each row show intermutational distance (IMD) of SNV in a single patient on chromosome 18. The BCL2 region is indicated by vertical red lines. (B-C) Mean SNV burden per Mb on chromosomes 11 and 14 in patients with MCL with a mutated IGHV region (M-IGHV) (B) and unmutated IGHV region (UM-IGHV) (C) on chromosome 14. The rightmost plot in each row shows IMD of SNV in a single patient from each group (M-IGHV or UM-IGHV) on chromosome 11. The CCND1 region is indicated by vertical red lines. (D-E) Mean SNV burden per Mb on chromosomes 11 and 14 in NDMM samples with t(11;14)(CCND1;IGH) in VDJ region and CSR region (D) and class-switch recombination (E). Rightmost plot in panels A-E shows IMD of SNV in a single patient from each group (VDJ or CSR) on chromosome 11. The CCND1 region is indicated by vertical red lines. (F) Proposed model behind the acquisition of CCND1 and BCL2 translocations in MM and B-cell lymphomas. (G) RAG RSS-motif analysis of SVs according to different categories as follows: CSR or VDJ region on chromosome 14 (RAG negative and positive controls, respectively), pregain deletion, postgain deletion, t(11;14)(CCND1;IGH) on chromosome 14 VDJ region, t(11;14)(CCND1;IGH) on chromosome 14 CSR region, or somatic (all other SV).
Defining the germinal center origin of t(11;14)(CCND1;IGH) in MM. (A) Showing the mean SNV burden per megabase (Mb) on chromosomes 18 and 14 in DLBCL and FL samples with t(14;18)(IGH;BCL2). The rightmost plots in each row show intermutational distance (IMD) of SNV in a single patient on chromosome 18. The BCL2 region is indicated by vertical red lines. (B-C) Mean SNV burden per Mb on chromosomes 11 and 14 in patients with MCL with a mutated IGHV region (M-IGHV) (B) and unmutated IGHV region (UM-IGHV) (C) on chromosome 14. The rightmost plot in each row shows IMD of SNV in a single patient from each group (M-IGHV or UM-IGHV) on chromosome 11. The CCND1 region is indicated by vertical red lines. (D-E) Mean SNV burden per Mb on chromosomes 11 and 14 in NDMM samples with t(11;14)(CCND1;IGH) in VDJ region and CSR region (D) and class-switch recombination (E). Rightmost plot in panels A-E shows IMD of SNV in a single patient from each group (VDJ or CSR) on chromosome 11. The CCND1 region is indicated by vertical red lines. (F) Proposed model behind the acquisition of CCND1 and BCL2 translocations in MM and B-cell lymphomas. (G) RAG RSS-motif analysis of SVs according to different categories as follows: CSR or VDJ region on chromosome 14 (RAG negative and positive controls, respectively), pregain deletion, postgain deletion, t(11;14)(CCND1;IGH) on chromosome 14 VDJ region, t(11;14)(CCND1;IGH) on chromosome 14 CSR region, or somatic (all other SV).
Prevalence and biologic impact of deletion acquired after large chromosomal gains
After initiation, the MM life history is characterized by the subsequent acquisition and selection of numerous additional driver events, such as aneuploidies and SVs. As expected, the number of postgain deletions within large chromosomal gains was significantly higher than the pregain deletions among all HY/IgH-neg sample on HY chromosomes (Wilcoxon rank sum test, P < .0001) (supplemental Figure 6; supplemental Table 6). A total of 1117 postgain deletion events were observed in 380 of 1173 (32.4%) samples from 372 of 1150 (32.3%) patients (Figure 5). The median size of these deletions was 6 174 100 bp (range, 8300-98 499 700). To investigate the biologic impact of these events, we restricted our analysis to 872 of 1117 (78.1%) postgain deletion events from 287 of 380 (75.5%) samples with available WGS and RNAseq data. SV-based postgain deletion events emerged as being involved in both the downregulation of TSGs (47/872 [5.4%] events in 39/287 [13.6%] samples) and the upregulation of oncogenes (25/872 [2.9%] events in 25/287 [8.7%] samples) (Figures 5A and 6; supplemental Table 6). The upregulation of oncogenes and the suppression of TSGs were significantly more pronounced than the expression levels observed in diploid regions unaffected by postgain deletions (P < .0001 and P < .0001, respectively; supplemental Figure 7). Furthermore, we identified 26 overexpressed fusion genes that were caused by these postgain deletions, 2 of which involved known oncodrivers, including PDCD1LG2 and MDM4 (Figure 6B; supplemental Figure 8; supplemental Table 7).
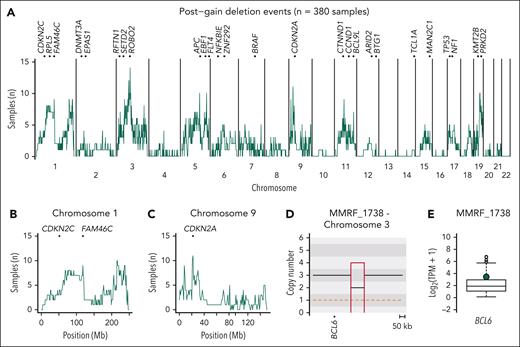
Postgain deletion prevalence and functional impact in patients with newly diagnosed and relapsed RM. (A) Cumulative distribution plot of 382 MM samples with postgain deletions. Top of the plot shows TSGs and oncogenes that were enriched with more than the median number of patients per 1 Mb bin having a postgain deletion on the given chromosome. (B) Enlarged distribution of postgain deletions per 1 Mb bin on chromosome 1 with involvement of the TSGs CDKN2C and FAM46C. (C) Enlarged distribution of postgain deletions per 1 Mb bin on chromosome 9 with involvement of the TSG CDKN2A. (D) CNV plot showing a postgain deletion near BCL6 in NDMM patient MMRF_1738. (E) RNAseq transcript levels of NDMM patients from the MMRF study with patient MMRF_1738 highlighted in green, showing overexpression.
Postgain deletion prevalence and functional impact in patients with newly diagnosed and relapsed RM. (A) Cumulative distribution plot of 382 MM samples with postgain deletions. Top of the plot shows TSGs and oncogenes that were enriched with more than the median number of patients per 1 Mb bin having a postgain deletion on the given chromosome. (B) Enlarged distribution of postgain deletions per 1 Mb bin on chromosome 1 with involvement of the TSGs CDKN2C and FAM46C. (C) Enlarged distribution of postgain deletions per 1 Mb bin on chromosome 9 with involvement of the TSG CDKN2A. (D) CNV plot showing a postgain deletion near BCL6 in NDMM patient MMRF_1738. (E) RNAseq transcript levels of NDMM patients from the MMRF study with patient MMRF_1738 highlighted in green, showing overexpression.
Functional impact of postgain deletions. Left column: copy-number plot. Right column: bulk RNAseq transcript for each involved gene. The green dot denotes transcript levels for the patients represented in copy-number plots. (A) Postgain deletion near the oncogene BCL2 led to a GOF. (B) Postgain deletion led to a gene fusion. (C) Postgain deletion involving TSG ATM led to gene expression downregulation (ie, LOF) despite having 2 remaining alleles. GOF, gain-of-function.
Functional impact of postgain deletions. Left column: copy-number plot. Right column: bulk RNAseq transcript for each involved gene. The green dot denotes transcript levels for the patients represented in copy-number plots. (A) Postgain deletion near the oncogene BCL2 led to a GOF. (B) Postgain deletion led to a gene fusion. (C) Postgain deletion involving TSG ATM led to gene expression downregulation (ie, LOF) despite having 2 remaining alleles. GOF, gain-of-function.
Early evidence of deletions capable of inducing oncogene gain-of-function in MM has been reported previously (eg, NSMCE2)28,44 and aligns with biologic expectations of oncogenes joint near a distal enhancer. However, the impact of deletions that reduce the copies of TSGs from 3 to 2 is a concept not firmly established in cancer. In fact, without integration of SV and CNV data, these events would be categorized as diploid and considered neutral. However, 95.7% (45/47) of these LOF events led to TSG downregulation that was comparable with monoallelic and biallelic deletions despite the presence of 2 heterozygous and intact alleles (Figure 6C). With the hypothesis that the downregulation of TSGs after postgain deletion was driven by gene-silencing mechanisms, we integrated CNV, SV, RNAseq, and scATACseq data from 8 patients with RRMM with a total of 13 samples (supplemental Methods).38 In 6 patients, a total of 17 postgain deletions were detected (supplemental Table 6). Based on RNAseq data, 4 of 17 postgain deletions led to an over- or underexpression of 11 known COSMIC driver genes when compared with the other RRMM samples (supplemental Figure 9A; supplemental Tables 6 and 8). A similar trend was seen at the chromatin accessibility level for 7 of 11 genes (supplemental Figure 9B; supplemental Table 8). One example is patient RRMM21 who acquired a postgain deletion at chr11q12-q13.4 between the 2 available time points, which led to a decrease in chromatin accessibility of CTNND1 (log2 fold-change T2/T1 [LFC] = −0.59), FEN1 (LFC = −0.46), MALAT1 (LFC = −0.66), and SDHAF2 (LFC = −0.69) at time point 2 (Figure 7; supplemental Figure 9C-F; supplemental Table 8). Especially for the 3 TSGs FEN1, MALAT1, and SDHAF2, the chromatin accessibility of the promotor and gene body was in the first-quartile of all investigated RRMM samples (Figure 7; supplemental Figure 9; supplemental Table 8), suggesting that downregulation at the expression and accessibility level of the TSGs was not just because of a copy number effect. Notably, a high number of postgain events (≥3) was associated with poor outcome when considering all patients (OS: P = .047; PFS: P = .0014, log-rank test) and HY/IgH-neg patients with NDMM only (OS: P = .016; PFS: P = .0075, log-rank test) (supplemental Figure 10). The prognostic impact was confirmed when considering the number of postgain deletions as a linear feature and was independent of high-risk cytogenetics [ie, TP53 mutations, TP53 deletions, t(4;14)(NSD2:IGH), t(14;16)(IGH:MAF), t(14;20)(IGH:MAFB)] and ISS staging, most likely reflecting a higher level of genomic complexity (supplemental Tables 9-12). In fact, in line with this observation, patients classified within the complex category, according to the recently proposed 12-groups classification,25 were enriched for postgain deletion events when compared with those who presented a simple genomic profile (supplemental Figure 11). Overall, because of the WGS resolution, we were able to define postgain deletions as a new protumor mechanism involved in gain-of-function and LOF events by affecting both chromatin accessibility and gene expression of the respective genes.
Single-cell ATACseq data from patients with RRMM reveal a novel mechanism of tumor suppressor inactivation. (A) Patient RRMM21 showed postgain deletion selected between 2 distinct time points (T1, the earliest; T2, the latest), which led to downregulation of 2 tumor suppressors, namely CTNND1 and MALAT1 (ie, loss of function). (B-C) Single-cell ATAC sequencing showed loss of chromatin accessibility after the postgain deletion near both genes between the 2 samples.
Single-cell ATACseq data from patients with RRMM reveal a novel mechanism of tumor suppressor inactivation. (A) Patient RRMM21 showed postgain deletion selected between 2 distinct time points (T1, the earliest; T2, the latest), which led to downregulation of 2 tumor suppressors, namely CTNND1 and MALAT1 (ie, loss of function). (B-C) Single-cell ATAC sequencing showed loss of chromatin accessibility after the postgain deletion near both genes between the 2 samples.
Discussion
In this study, we conducted a comprehensive investigation using high-coverage WGS resolution to define the genomic events and mechanisms involved in the early phases of MM pathogenesis. Our innovative analytical approach that integrated molecular time, SV, CNV, and RNAseq data revealed the existence of genomic events that were acquired before trisomies in HY patients, a previously presumed initiating event in MM for >30 years.1,5,7 Notably, the significant impact of these events on both TSGs and oncogenes supports their potential role as drivers. In contrast with normal B-cell deletions,20 these events are likely acquired between when B cells exit the BM and the GC where HY is known to occur16,26 and not by off-target RAG activity in the BM. Although we can exclude RAG’s primary role in the acquisition of VDJ t(11;14)(CCND1;IGH), we cannot rule out the possibility of rare SVs mediated by RAG, akin to those observed in normal B cells.20 Overall, data generated by our analytical workflow demonstrated that RAG plays either a null or marginal role in MM pathogenesis, thereby resolving a decade-long debate. In addition, the existence of events acquired before HY in IgH-neg patients significantly reshapes our historic model of MM pathogenesis, expanding its heterogeneity and complexity.
In our investigation of SVs within large chromosomal gains, we observed a high prevalence of postgain deletions that led to both LOF and gain-of-function events. Gain-of-function events that are mediated by postgain deletions are not extensively studied in cancer but are not unknown. In contrast, to the best of our knowledge, deletions that occur after the gain that induces a LOF in a TSG, despite the diploid copy number status, represent a potential new mechanism with implications for other cancers, particularly those with large gains and whole-genome doubling.31,45 Importantly, these events would be missed by targeted/exome approaches and would not be included in tools aimed at identifying recurrent deletions across cancer genomes (eg, GISTIC [Genomic Identification of Significant Targets in Cancer]). Although our WGS NDMM series is one of the largest published so far, we recognize that a larger data set would be needed to test whether the prognostic impact of postgain losses of individual driver genes is similar to that observed in standard deletions.
Overall, because of their prevalence, clinical impact, and involvement of key oncogenes and TSGs, we propose that pre- and post-large chromosomal gain deletions represent novel mechanisms that contribute to the acquisition of driver events in the intricate pathogenesis and life history of MM.
Acknowledgments
The Heidelberg team thanks the Sample Processing Lab, the High Throughput Sequencing unit of the Genomics and Proteomics Core Facility and the Omics IT and Data Management Core Facility of the German Cancer Research Center (DKFZ), the DKFZ-Heidelberg Center for Personalized Oncology office, the Biobank Multiple Myeloma GMMG Central Lab and Biobank Multiple Myeloma, University Hospital Heidelberg, and the Myeloma Registry for excellent services. CoMMpass data were generated by the Multiple Myeloma Research Foundation.
This work was supported by the Myeloma Solutions Fund, the Paula and Rodger Riney Multiple Myeloma Research Program Fund, the Tow Foundation, a Sylvester Comprehensive Cancer Center National Institutes of Health (NIH), National Cancer Institute (NCI) Core Grant (P30 CA 240139), a Memorial Sloan Kettering Cancer Center NIH, NCI Core Grant (P30 CA 008748), and a New York University NIH, NCI Core Grant (P30CA016087). F.M. is supported by the Leukemia and Lymphoma Society and by the International Myeloma Society. G.J.M. received grant support through a Translational Research Program award from the Leukemia and Lymphoma Society (6020-20). K.M. received funding from the Multiple Myeloma Research Foundation, the American Society of Hematology, and the International Myeloma Society. A.M.P. is funded by the Medical Data Scientist Program of the Heidelberg University Faculty of Medicine. Support and funding of the project via the Dietmar Hopp Foundation and the NCT Heidelberg Molecular Precision Oncology Program (project K08K) are gratefully acknowledged. This publication was supported through state funds approved by the State Parliament of Baden-Württemberg for the Innovation Campus Health + Life Science Alliance Heidelberg Mannheim. Data storage service via SDS@hd was supported by the Ministry of Science, Research and the Arts Baden-Württemberg and the German Research Foundation through grants INST 35/1314-1 FUGG and INST 35/1503-1 FUGG.
Authorship
Contribution: F.M., M.S.R., and N.W. designed and supervised the study, collected, generated, and analyzed the data and wrote the manuscript; B.Z., A.M.C., and A.M.P. collected, generated, analyzed the data, and wrote the manuscript; M.K. analyzed the data; N.K., S.U., O.L., E.K.M., H.G., K.C.W., A.M.L., and R.F. supervised the clinical trials, collected data; and K.M., K.R., M.C., B.D., P.B., L.J., P.R., S.H., M.-A.B., D.G., Y.Z., F.D., G.J.M., and A.D. collected the data.
Conflict-of-interest disclosure: O.L. reports receiving research funding from the NIH NCI, the US Food and Drug Administration, the Multiple Myeloma Research Foundation, the International Myeloma Foundation, the Leukemia and Lymphoma Society, the Myeloma Solutions Fund, the Paula and Rodger Riney Multiple Myeloma Research Program Fund, the Tow Foundation, the Perelman Family Foundation, the Rising Tide Foundation, Amgen, Celgene, Janssen, Takeda, Glenmark, Seattle Genetics, and Karyopharm; receiving honoraria from or serving on advisory boards of Adaptive, Amgen, Binding Site, Bristol Myers Squibb (BMS), Celgene, Cellectis, Glenmark, Janssen, Juno, and Pfizer; and serving on the independent data monitoring committees for clinical trials lead by Takeda, Merck, Janssen, and Theradex. G.J.M. reports receiving funding from the NIH NCI, the Multiple Myeloma Research Foundation, the Leukemia and Lymphoma Society, the Perelman Family Foundation, Amgen, Celgene, Janssen, and Takeda; receiving honoraria or serving on advisory boards for Adaptive, Amgen, BMS, Celgene, and Janssen; and serving on the independent data monitoring committees for clinical trials led by Takeda, Karyopharm, and Sanofi. E.K.M. reports serving in a consulting or advisory role for and receiving honoraria, research funding, travel accommodation, and expenses from BMS/Celgene, GlaxoSmithKline, Janssen-Cilag, Sanofi, Stemline, and Takeda. K.C.W. reports receiving research grants from AbbVie, Amgen, BMS/Celgene, Janssen, GlaxoSmithKline, and Sanofi and receiving honoraria and consulting fees from AbbVie, Amgen, Adaptive Biotech, AstraZeneca, BMS/Celgene, BeiGene, Janssen, GlaxoSmithKline, Karyopharm, Novartis, Oncopeptides, Pfizer, Roche Pharma, Sanofi, Stemline, and Takeda. R.F. reports serving in a consulting or advisory role for and receiving honoraria, travel accommodation, and expenses from Amgen, BMS/Celgene, GlaxoSmithKline, Janssen-Cilag, Sanofi, Stemline, and Takeda. H.G. reports receiving grants and/or investigational medicinal products from Amgen, Array Biopharma/Pfizer, BMS/Celgene, Chugai, the Dietmar Hopp Foundation, Janssen, Johns Hopkins University, Mundipharma GmbH, and Sanofi; receiving research support from Amgen, BMS, Celgene, GlycoMimetics Inc, GlaxoSmithKline, Heidelberg Pharma, Hoffmann-La Roche, Karyopharm, Janssen, Incyte Corporation, Millennium Pharmaceuticals Inc, Molecular Partners, Merck Sharp and Dohme, MorphoSys AG, Pfizer, Sanofi, Takeda, and Novartis; serving on advisory boards for Amgen, BMS, Janssen, Sanofi, and Adaptive Biotechnology; receiving honoraria from Amgen, BMS, Chugai, GlaxoSmithKline, Janssen, Novartis, Sanofi, and Pfizer; and receiving support for attending meetings and/or travel from Amgen, BMS, GlaxoSmithKline, Janssen, Novartis, Sanofi, and Pfizer. F.M. reports serving as a consultant for Medidata. The remaining authors declare no competing financial interests.
Correspondence: Francesco Maura, Division of Myeloma, Department of Medicine, Sylvester Myeloma Institute Sylvester Comprehensive Cancer Center, University of Miami Don Soffer Clinical Research Center, 1120 NW 14th St, Clinical Research Building, Miami, FL 33136; email: fxm557@med.miami.edu; and Niels Weinhold, Heidelberg Myeloma Center, Department of Medicine V, University Hospital Heidelberg, Im Neuenheimer Feld 410, 69120 Heidelberg, Germany; email: niels.weinhold@med.uni-heidelberg.de.
References
Author notes
A.M.C., A.M.P., and B.Z. contributed equally to this work.
M.S.R., N.W., and F.M. jointly supervised this work.
German-Speaking Myeloma Multicenter Group HD6 trial (GMMG-HD6) data are currently in the process of being uploaded on European Genome-phenome Archive (EGA): EGAS00001007469. MSKCC WGS data have been uploaded on EGA: EGAD00001011132. RRMM WGS, RNAseq and single-cell ATACseq data have been uploaded on EGA: EGAS00001006538, EGAS00001004363, EGAS00001004805 and EGAS00001005973. CoMMpass, MCL, and Pan-Cancer Analysis of Whole Genomes (PCAWG) data are available on phs000748, https://dcc.icgc.org/pcawg/, and EGAS00001004165, respectively.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal