Key Points
Donor age was a more consistently influential factor as a main effect, with a nonlinear relationship with overall mortality hazard.
The influence of donor type may be more complex, potentially becoming more pronounced in specific contexts, such as with older donors.
Visual Abstract
Limited data exist comparing haploidentical and mismatched unrelated donor (MMUD) hematopoietic cell transplantation (HCT) with posttransplantation cyclophosphamide for graft-versus-host disease prophylaxis, especially considering donor age. Herein, we report the outcomes of 660 haploidentical and 195 MMUD HCT recipients treated at MD Anderson Cancer Center. Beyond standard Cox proportional hazards modeling, we used inverse probability of treatment weighting (IPTW) and matched-pair analysis, and performed additional analysis by incorporating an external MMUD validation cohort from the Center for International Blood and Marrow Transplant Research (CIBMTR). The primary outcome was overall survival (OS). In multivariable analysis, haploidentical donors had a hazard ratio (HR) of 1.20 (95% confidence interval [CI], 0.93-1.54; P = .16) compared with the MMUD group. Donor age showed a nonlinear association with OS. These findings were corroborated by IPTW, matched-pair analyses, and CIBMTR validation analyses. Exploratory analysis revealed inferior OS for older (age of >50 years) haploidentical donor group compared with younger (age of <30 years) MMUD recipients (HR, 1.91; 95% CI, 1.21-3.01; P = .005). Our analyses suggest that although donor type may play a role, there was a more prominent role for donor age in influencing OS. Moreover, our findings indicate a potential nuance wherein the impact of donor type may vary by donor age. Further research, particularly with larger cohorts, is needed to fully elucidate the complex and potentially interacting roles of donor type and donor age, along with HLA factors.
Introduction
A significant proportion of patients requiring allogeneic hematopoietic cell transplantation (HCT) lack access to HLA-matched donors, with estimates ranging between 16% to 52% among patients from ethnically diverse backgrounds.1,2 This highlights the importance of exploring alternative donor options, with haploidentical donors being the most common choice, followed by HLA-mismatched unrelated donors (MMUD).3 The use of MMUDs was traditionally limited by the increased risk of graft-versus-host disease (GVHD), graft failure, and infections reported with calcineurin inhibitor–based GVHD.4,5 However, the significantly improved outcomes with the use of posttransplantation cyclophosphamide (PTCy) prophylaxis6-8 made the results of MMUD HCT comparable with those of matched unrelated donor HCT.9
Two studies from the European Group for Blood and Marrow Transplantation registry compared the outcomes of haploidentical and MMUD HCT with PTCy for GVHD prophylaxis.10,11 However, donor age, a significant predictor of survival,12-15 was not included in these analyses. Our study aims to compare the outcomes after haploidentical and MMUD HCT with PTCy for GVHD prophylaxis while accounting for donor age and other relevant non-HLA characteristics. We hypothesized that, in the setting of PTCy-based GVHD prophylaxis that attenuates HLA disparity,9,16 donor type would not be an independent predictor of overall survival (OS), and that donor age would emerge as a prognostic marker in patients undergoing mismatched HCT.
Methods
We included all consecutive adult patients with any hematologic malignancy who underwent their first allogeneic HCT with either a haploidentical donor (n = 660) or MMUD (n = 195) with PTCy prophylaxis at MD Anderson Cancer Center (MDACC) from June 2008 to August 2024. Additional validation analysis was performed using a publicly available Center for International Blood and Marrow Transplant Research (CIBMTR) data set17 from the referenced publication9 that included patients who underwent MMUD HCT with PTCy (supplemental Figure 1). Haploidentical donors were defined as related donors with ≥2 HLA mismatches in the nonshared haplotype. Unrelated donors with ≤9 of 10 HLA matches (MDACC cohort) or ≤7 of 8 HLA matches (CIBMTR cohort) were classified as MMUDs. Although high-resolution HLA typing was performed for all patients, detailed HLA data were not available for this analysis. The primary outcome of interest was OS. Secondary outcomes included relapse, nonrelapse mortality (NRM), grade 3 to 4 acute GVHD (aGVHD), and any chronic GVHD (cGVHD). The local institutional review board (number 2024-0040) approved the study, all participants included in this study provided written informed consent, and the study was conducted in accordance with the Declaration of Helsinki.
Statistical analysis
We used a multifaceted analytical approach to compare outcomes after haploidentical and MMUD HCT. First, conventional Cox proportional hazards (PH) multivariable analysis (MVA) was conducted. However, notable differences in baseline characteristics were observed between the 2 donor groups, raising concerns that the results of the Cox PH model might be influenced by these imbalances. Therefore, to address these concerns, we implemented 2 additional strategies: inverse probability of treatment weighting (IPTW) analysis and propensity score–matched analysis. The methods for these analyses are elaborated hereafter.
Univariate analyses
In univariate analyses, OS probability was estimated using the Kaplan-Meier method. The cumulative incidence method, accounting for competing risks, was used to estimate the probabilities of relapse, NRM, aGVHD, and cGVHD. Competing risks considered were disease relapse for NRM, death without relapse (NRM) for relapse, and death for GVHD. Median follow-up time was calculated using the reverse Kaplan-Meier method.
Cox PH models
Multivariable analyses were performed using Cox PH models on cause-specific hazards for all outcomes with competing events. All variables listed in Table 1 were included in the univariate analyses (supplemental Tables 1 and 2). Variables with P values <.10 in univariate analyses were included in the multivariable models. Then, stepwise selection with both forward and backward selection was used to identify a more parsimonious model retaining all significant variables with P value ≤.05. However, all donor-related factors, including donor type, donor age, donor-recipient sex, and donor-recipient cytomegalovirus (CMV) status, were retained in the final multivariate models regardless of their statistical significance. The PH assumption was assessed for all variables graphically (log-log plots, Schoenfeld residuals) and by including time interactions in the model. Variables violating the PH assumption were adjusted through stratification.
Baseline characteristics
| . | Overall population . | Matched-pair cohort . | ||
|---|---|---|---|---|
| Haplo (n = 660) . | MMUD (n = 195) . | Haplo (n = 327) . | MMUD (n = 195) . | |
| Recipient age, median (Q1-Q3), y | 52 (37-61) | 56 (44-65) | 55 (41-62) | 56 (44-65) |
| Recipient age, n (%) | ||||
| <50 years | 315 (47.7) | 72 (36.9) | 131 (40.1) | 72 (36.9) |
| ≥50 years | 345 (52.3) | 123 (63.1) | 196 (59.9) | 123 (63.1) |
| Donor age, median (Q1-Q3), y | 34 (24.8-43) | 33 (26-41.5) | 33 (24-43) | 33 (26-41.5) |
| Donor age, y, n (%) | ||||
| <30 years | 271 (41.1) | 83 (43.7) | 142 (43.4) | 83 (43.7) |
| ≥30 years | 389 (58.9) | 107 (56.3) | 185 (56.6) | 107 (56.3) |
| Disease, n (%) | ||||
| AML/MDS | 385 (58.3) | 119 (61.0) | 176 (53.8) | 119 (61.0) |
| ALL | 99 (15.0) | 14 (7.2) | 38 (11.6) | 14 (7.2) |
| Other∗ | 176 (26.7) | 62 (31.8) | 113 (34.6) | 62 (31.8) |
| Disease stage, n (%) | ||||
| Low | 50 (7.6) | 21 (10.8) | 33 (10.1) | 21 (10.8) |
| Intermediate | 372 (56.4) | 107 (54.9) | 200 (61.2) | 107 (54.9) |
| High | 197 (29.8) | 47 (24.1) | 77 (23.5) | 47 (24.1) |
| Very high | 32 (4.8) | 10 (5.1) | 8 (2.4) | 10 (5.1) |
| Missing | 9 (1.4) | 10 (5.1) | 9 (2.8) | 10 (5.1) |
| Conditioning intensity, n (%) | ||||
| MAC | 410 (62.1) | 140 (71.8) | 217 (66.4) | 140 (71.8) |
| RIC/NMA | 250 (37.9) | 55 (28.2) | 110 (33.6) | 55 (28.2) |
| Graft type, n (%) | ||||
| Bone marrow | 466 (70.6) | 71 (36.4) | 142 (43.4) | 71 (36.4) |
| Peripheral blood | 194 (29.4) | 124 (63.6) | 185 (56.6) | 124 (63.6) |
| HCT-CI, n (%) | ||||
| 0 | 120 (18.2) | 42 (21.5) | 66 (20.2) | 42 (21.5) |
| 1-2 | 191 (28.9) | 49 (25.1) | 80 (24.5) | 49 (25.1) |
| ≥3 | 349 (52.9) | 104 (53.3) | 181 (55.4) | 104 (53.3) |
| D/R CMV, n (%) | ||||
| Neg/Neg | 84 (12.8) | 32 (16.5) | 57 (17.4) | 32 (16.4) |
| Neg/Pos | 196 (29.9) | 72 (37.1) | 92 (28.1) | 72 (36.9) |
| Pos/Neg | 52 (7.9) | 25 (12.9) | 27 (8.3) | 25 (12.8) |
| Pos/Pos | 324 (49.4) | 65 (33.5) | 151 (46.2) | 66 (33.8) |
| D/R sex, n (%) | ||||
| Female to male | 138 (20.9) | 41 (21.2) | 63 (19.3) | 41 (21.0) |
| Not female to male | 522 (79.1) | 152 (78.8) | 264 (80.7) | 154 (79.0) |
| KPS, n (%) | ||||
| <90 | 226 (34.2) | 84 (43.1) | 117 (35.8) | 84 (43.1) |
| ≥90 | 323 (48.9) | 88 (45.1) | 157 (48.0) | 88 (45.1) |
| Missing | 111 (16.8) | 23 (11.8) | 53 (16.2) | 23 (11.8) |
| Race, n (%) | ||||
| White | 358 (54.2) | 140 (71.8) | 192 (58.7) | 140 (71.8) |
| Others | 294 (44.5) | 53 (27.2) | 132 (40.4) | 53 (27.2) |
| Missing | 8 (1.2) | 2 (1.0) | 3 (0.9) | 2 (1.0) |
| Time to HCT, n (%) | ||||
| <6 months | 180 (27.4) | 45 (23.1) | 73 (22.3) | 45 (23.1) |
| 6-12 months | 165 (25.1) | 61 (31.3) | 94 (28.7) | 61 (31.3) |
| >12 months | 313 (47.6) | 89 (45.6) | 160 (48.9) | 89 (45.6) |
| Year of HCT, n (%) | ||||
| <2019 | 324 (49.1) | 69 (35.4) | 121 (37.0) | 69 (35.4) |
| ≥2019 | 336 (50.9) | 126 (64.6) | 206 (63.0) | 126 (64.6) |
| Follow-up, median (Q1-Q3), mo† | 54.20 (30.32-84.79) | 25.26 (13.60-62.29) | 49.9 (29.40-76.58) | 25.26 (13.60-62.29) |
| . | Overall population . | Matched-pair cohort . | ||
|---|---|---|---|---|
| Haplo (n = 660) . | MMUD (n = 195) . | Haplo (n = 327) . | MMUD (n = 195) . | |
| Recipient age, median (Q1-Q3), y | 52 (37-61) | 56 (44-65) | 55 (41-62) | 56 (44-65) |
| Recipient age, n (%) | ||||
| <50 years | 315 (47.7) | 72 (36.9) | 131 (40.1) | 72 (36.9) |
| ≥50 years | 345 (52.3) | 123 (63.1) | 196 (59.9) | 123 (63.1) |
| Donor age, median (Q1-Q3), y | 34 (24.8-43) | 33 (26-41.5) | 33 (24-43) | 33 (26-41.5) |
| Donor age, y, n (%) | ||||
| <30 years | 271 (41.1) | 83 (43.7) | 142 (43.4) | 83 (43.7) |
| ≥30 years | 389 (58.9) | 107 (56.3) | 185 (56.6) | 107 (56.3) |
| Disease, n (%) | ||||
| AML/MDS | 385 (58.3) | 119 (61.0) | 176 (53.8) | 119 (61.0) |
| ALL | 99 (15.0) | 14 (7.2) | 38 (11.6) | 14 (7.2) |
| Other∗ | 176 (26.7) | 62 (31.8) | 113 (34.6) | 62 (31.8) |
| Disease stage, n (%) | ||||
| Low | 50 (7.6) | 21 (10.8) | 33 (10.1) | 21 (10.8) |
| Intermediate | 372 (56.4) | 107 (54.9) | 200 (61.2) | 107 (54.9) |
| High | 197 (29.8) | 47 (24.1) | 77 (23.5) | 47 (24.1) |
| Very high | 32 (4.8) | 10 (5.1) | 8 (2.4) | 10 (5.1) |
| Missing | 9 (1.4) | 10 (5.1) | 9 (2.8) | 10 (5.1) |
| Conditioning intensity, n (%) | ||||
| MAC | 410 (62.1) | 140 (71.8) | 217 (66.4) | 140 (71.8) |
| RIC/NMA | 250 (37.9) | 55 (28.2) | 110 (33.6) | 55 (28.2) |
| Graft type, n (%) | ||||
| Bone marrow | 466 (70.6) | 71 (36.4) | 142 (43.4) | 71 (36.4) |
| Peripheral blood | 194 (29.4) | 124 (63.6) | 185 (56.6) | 124 (63.6) |
| HCT-CI, n (%) | ||||
| 0 | 120 (18.2) | 42 (21.5) | 66 (20.2) | 42 (21.5) |
| 1-2 | 191 (28.9) | 49 (25.1) | 80 (24.5) | 49 (25.1) |
| ≥3 | 349 (52.9) | 104 (53.3) | 181 (55.4) | 104 (53.3) |
| D/R CMV, n (%) | ||||
| Neg/Neg | 84 (12.8) | 32 (16.5) | 57 (17.4) | 32 (16.4) |
| Neg/Pos | 196 (29.9) | 72 (37.1) | 92 (28.1) | 72 (36.9) |
| Pos/Neg | 52 (7.9) | 25 (12.9) | 27 (8.3) | 25 (12.8) |
| Pos/Pos | 324 (49.4) | 65 (33.5) | 151 (46.2) | 66 (33.8) |
| D/R sex, n (%) | ||||
| Female to male | 138 (20.9) | 41 (21.2) | 63 (19.3) | 41 (21.0) |
| Not female to male | 522 (79.1) | 152 (78.8) | 264 (80.7) | 154 (79.0) |
| KPS, n (%) | ||||
| <90 | 226 (34.2) | 84 (43.1) | 117 (35.8) | 84 (43.1) |
| ≥90 | 323 (48.9) | 88 (45.1) | 157 (48.0) | 88 (45.1) |
| Missing | 111 (16.8) | 23 (11.8) | 53 (16.2) | 23 (11.8) |
| Race, n (%) | ||||
| White | 358 (54.2) | 140 (71.8) | 192 (58.7) | 140 (71.8) |
| Others | 294 (44.5) | 53 (27.2) | 132 (40.4) | 53 (27.2) |
| Missing | 8 (1.2) | 2 (1.0) | 3 (0.9) | 2 (1.0) |
| Time to HCT, n (%) | ||||
| <6 months | 180 (27.4) | 45 (23.1) | 73 (22.3) | 45 (23.1) |
| 6-12 months | 165 (25.1) | 61 (31.3) | 94 (28.7) | 61 (31.3) |
| >12 months | 313 (47.6) | 89 (45.6) | 160 (48.9) | 89 (45.6) |
| Year of HCT, n (%) | ||||
| <2019 | 324 (49.1) | 69 (35.4) | 121 (37.0) | 69 (35.4) |
| ≥2019 | 336 (50.9) | 126 (64.6) | 206 (63.0) | 126 (64.6) |
| Follow-up, median (Q1-Q3), mo† | 54.20 (30.32-84.79) | 25.26 (13.60-62.29) | 49.9 (29.40-76.58) | 25.26 (13.60-62.29) |
ALL, acute lymphoblastic leukemia; D/R, donor/recipient; Haplo, haploidentical; KPS, Karnofsky performance status; MAC, myeloablative conditioning; Neg, negative; NMA, nonmyeloablative conditioning; Pos, positive; Q1, first quartile; Q3, third quartile; RIC, reduced-intensity conditioning.
Other: chronic lymphocytic leukemia, chronic myeloid leukemia, lymphoma, other hematologic malignancies.
Follow-up was calculated using the inverse Kaplan-Meier method.
IPTW analysis
For IPTW analysis, a covariate balancing propensity score (CBPS) model was used to estimate propensity scores while optimizing covariate balance using the CBPS package in R.18 The logistic regression model for propensity score estimation included the following clinically relevant baseline covariates: recipient age, donor age, graft type, conditioning intensity, disease, disease risk index (DRI), and HCT-specific comorbidity index (HCT-CI). Weights derived from the CBPS model were then applied to conduct the IPTW analysis. IPTW is a statistical method that adjusts for confounding variables by assigning weights to each observation based on the probability of receiving the treatment (donor) they actually received. This created a pseudopopulation in which the distribution of baseline characteristics was comparable between the haploidentical and MMUD groups. This approach minimizes the influence of confounding factors on the observed treatment effects, allowing for a more accurate and unbiased comparison of outcomes. Covariate balance was assessed using standardized mean differences and visualized with Love plots (supplemental Figure 2).
Matched-pair analysis
To generate a matched-pair cohort, a 1:2 nearest-neighbor matching without replacement was performed using the MatchIt package.19 As with the IPTW analysis, propensity scores for matching were derived from a logistic regression model including recipient age, donor age, graft type, conditioning intensity, disease, DRI, and HCT-CI. This matching procedure aimed to create groups that were more comparable with respect to these baseline characteristics. In 1:2 matching, each MMUD recipient was matched to up to 2 of the closest haploidentical patients based on propensity scores, without replacement. Therefore, some MMUD patients may have been successfully matched to 2 unique haploidentical patients. Other MMUD patients, depending on the distribution of propensity scores and the constraints of exact matching, may have been matched to 1 haploidentical patient. After matching, Cox PH MVA was conducted on the matched-pair cohort. In these models, the matching is accounted for in the study design by analyzing outcomes within the balanced matched cohort. The Cox PH MVA on the matched pairs served to assess the donor type effect within this balanced cohort and to further adjust for any residual confounding or other relevant covariates not perfectly balanced by matching.
Modeling donor age
Because donor age was a key variable of interest, we first explored its functional form and tested the linearity assumption. Initially, a natural spline model was fitted with donor age in the univariate analysis, revealing significant nonlinear effects for the spline terms (supplemental Figure 3). Therefore, donor age was modeled in 2 primary ways. First, donor age was categorized into 3 groups: age of <30 years, 30 to 50 years, and >50 years, using data-driven cutoff points identified by maximally selected rank statistics as ∼26 years for MMUD and 50 years for haploidentical donors. However, categorization introduces the risk of losing important information through arbitrary cutoffs, potentially oversimplifying the data and ignoring subtle variations within each category. Therefore, donor age was also modeled as spline term using restricted cubic splines with 4 degrees of freedom to capture the nonlinear relationship and avoid arbitrary categorization. Considering the advantages and disadvantages of each approach, we present the results of both models: donor age as splines as well as illustrative categorical representations.
Validation analysis using an external CIBMTR cohort
Lastly, an analysis was performed using an external validation cohort from an existing CIBMTR data set,9 which included patients who underwent MMUD HCT with PTCy prophylaxis (MMUD-CIBMTR). A similar cohort for haploidentical donors from the CIBMTR was not publicly available; therefore, MMUD-CIBMTR patients were compared with those who underwent haploidentical HCT at MDACC. This analysis was restricted to only patients with acute leukemia who underwent HCT within, or after, 2017, to minimize heterogeneity. These patients were matched to account for baseline differences between the groups using a propensity score matching approach. Propensity scores were calculated using the MatchIt package, with donor type as the dependent variable. To partly mitigate the impact of potential practice variations across centers, we used exact matching on key clinical variables (graft type, donor age, disease, DRI, conditioning, HCT-CI, and Karnofsky performance status) while incorporating recipient age and sex as additional covariates. The nearest-neighbor matching method was applied with a ratio of 1:1 to create comparable groups. The matched data set was subsequently used for further statistical analyses including Cox PH multivariate modeling, ensuring that the impact of confounding variables was minimized, and the findings were reflective of true associations. Then, Cox PH MVA was conducted on the matched-pair cohort.
Assessment of interactions
Interaction effects between the main effects (donor type and donor age) and other statistically significant covariates were tested in all multivariate models. To assess the interaction between donor type and donor age in the categorical model, an interaction term between donor age (categorized into age of >50 years, 30-50 years, and <30 years) and donor type was included. In addition to assessing individual factor level interactions, the overall significance of this interaction was evaluated using a likelihood ratio test (LRT), comparing the interaction model with a reduced model without the interaction term. Similarly, to assess the effect of donor age on survival in the spline model, an interaction model was fitted. This model included an interaction term between the natural spline of donor age and donor type. The significance of this interaction was tested using a LRT, comparing the interaction model with a reduced model containing only the main effects. Interactions between donor age and all other significant variables in the model, and between donor type and all other significant variables in the model, were tested in a similar fashion.
All analyses were conducted using R Statistical Software (version 4.3.1; R Core Team 2023).
Results
The baseline characteristics were remarkably different between the donor groups in the overall MDACC cohort (haploidentical, n = 660; MMUD, n = 195), but the 2 groups were well-balanced in the matched-pair cohort (haploidentical, n = 327; MMUD, n = 195) as shown in Table 1. Specifically, the median age of patients was 55 and 56 years in the haploidentical and MMUD groups, respectively. In both groups, the median donor age was 33 years; acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) were the most common diseases (54%-61%), most patients had a DRI of intermediate or high and received myeloablative conditioning. The median follow-up in the haploidentical group was 54.2 months, and 25.3 months in the MMUD group.
OS
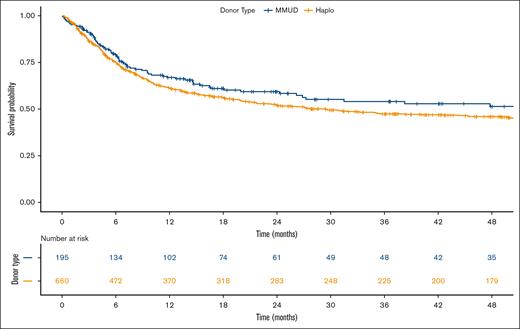
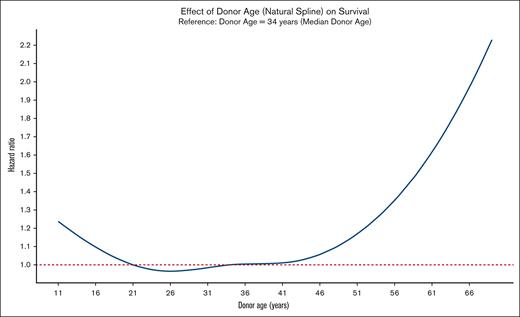
The Kaplan-Meier curves depicting OS for both donor types are presented in Figure 1. Multivariable analysis of overall mortality revealed the following hazard ratios (HRs) for haploidentical donors compared with MMUD: 1.20 (95% confidence interval [CI], 0.93-1.54; P = .16) in the Cox PH model, 1.22 (95% CI, 0.93-1.60; P = .15) in the IPTW analysis, and 1.15 (95% CI, 0.87-1.52; P = .32) in the matched-pair analysis. For donors aged >50 years, HRs for overall mortality were higher across all models: 1.46 (95% CI, 1.09-1.97; P = .01) in the Cox PH model, 1.52 (95% CI, 1.05-2.22; P = .02) in the IPTW analysis, and 1.68 (95% CI, 1.13-2.50; P = .01) in the matched-pair analysis. The results of MVA using donor age as spline function also revealed consistent patterns (Table 2). Figure 2 illustrates the nonlinear relationship between donor age and overall mortality hazard, as estimated by the Cox PH MVA model using a natural spline.
Kaplan-Meier estimate of OS comparing Haplo donor (yellow line) and MMUD (blue line) groups. Haplo, haploidentical.
Kaplan-Meier estimate of OS comparing Haplo donor (yellow line) and MMUD (blue line) groups. Haplo, haploidentical.
Multivariate analysis of outcomes: MDACC population
| . | N . | Events . | Cox PH (entire cohort) . | IPTW analysis (entire cohort) . | Cox PH (matched-pair cohort) . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HR . | 95% CI . | P value . | HR . | 95% CI . | P value . | HR . | 95% CI . | P value . | |||
| OS | |||||||||||
| Recipient age, y | |||||||||||
| <50 | 387 | 160 | Ref | ||||||||
| ≥50 | 468 | 265 | 1.70 | 1.39-2.09 | <.001 | 1.82 | 1.36-2.43 | <.001 | 1.85 | 1.38-2.47 | <.001 |
| Donor age, spline∗ | .05 | .049 | .05 | ||||||||
| Donor age, y∗ | .03 | .08 | .03 | ||||||||
| <30 | 354 | 163 | Ref | ||||||||
| 30-50 | 392 | 196 | 1.04 | 0.84-1.29 | .72 | 1.09 | 0.82-1.45 | .53 | 1.02 | 0.77-1.36 | .87 |
| >50 | 104 | 66 | 1.46 | 1.09-1.97 | .01 | 1.52 | 1.05-2.22 | .02 | 1.68 | 1.13-2.50 | .01 |
| Donor type | |||||||||||
| MMUD | 195 | 78 | Ref | ||||||||
| Haplo | 660 | 347 | 1.20 | 0.93-1.54 | .16 | 1.22 | 0.93-1.60 | .15 | 1.15 | 0.87-1.52 | .32 |
| Disease | .07 | ||||||||||
| ALL | 113 | 63 | Ref | ||||||||
| AML/MDS | 504 | 254 | - | - | - | 0.64 | 0.39-1.06 | .08 | - | - | - |
| Other | 238 | 108 | - | - | - | 0.88 | 0.51-1.51 | .64 | - | - | - |
| DRI | <.001 | <.001 | <.001 | ||||||||
| Low | 71 | 24 | Ref | ||||||||
| Intermediate | 479 | 201 | 1.23 | 0.80-1.90 | .35 | 1.48 | 0.80-2.72 | .21 | 1.19 | 0.70-2.00 | .51 |
| High | 244 | 154 | 2.13 | 1.37-3.31 | .001 | 2.77 | 1.46-5.26 | .002 | 1.88 | 1.09-3.24 | .02 |
| Very high | 42 | 36 | 4.20 | 2.48-7.11 | <.001 | 4.91 | 2.41-10.01 | <.001 | 4.84 | 2.39-9.78 | <.001 |
| Missing | 19 | 10 | 2.05 | 0.97-4.31 | .06 | 2.46 | 1.04-5.85 | .04 | 2.08 | 0.94-4.61 | .07 |
| HCT-CI | .002 | .04 | .11 | ||||||||
| 0 | 162 | 63 | Ref | ||||||||
| 1-2 | 240 | 107 | 1.00 | 0.73-1.37 | .99 | 0.89 | 0.60-1.30 | .54 | 0.87 | 0.58-1.31 | .50 |
| ≥3 | 453 | 255 | 1.42 | 1.07-1.88 | .01 | 1.28 | 0.91-1.81 | .16 | 1.22 | 0.86-1.73 | .26 |
| D/R CMV | .14 | .06 | .53 | ||||||||
| Neg/Neg | 117 | 48 | Ref | ||||||||
| Neg/Pos | 268 | 148 | 1.33 | 0.96-1.85 | .09 | 1.55 | 1.03-2.34 | .03 | 1.30 | 0.86-1.97 | .21 |
| Pos/Neg | 77 | 31 | 0.92 | 0.58-1.46 | .73 | 1.11 | 0.65-1.89 | .69 | 1.00 | 0.57-1.75 | .99 |
| Pos/Pos | 393 | 198 | 1.12 | 0.81-1.55 | .48 | 1.11 | 0.74-1.66 | .61 | 1.13 | 0.76-1.69 | .55 |
| D/R sex mismatch | |||||||||||
| F-to-M | 676 | 337 | Ref | ||||||||
| Not F-to-M | 179 | 88 | 0.94 | 0.74-1.19 | .61 | 0.92 | 0.69-1.24 | .58 | 0.86 | 0.62-1.18 | .35 |
| NRM† | |||||||||||
| Recipient age, y | |||||||||||
| <50 | 387 | 77 | Ref | ||||||||
| ≥50 | 468 | 169 | 1.78 | 1.44-2.20 | <.001 | 1.77 | 1.33-2.38 | <.001 | 2.25 | 1.52-3.33 | <.001 |
| Donor age, spline∗ | .036 | .02 | .06 | ||||||||
| Donor age, y∗ | .02 | .06 | .009 | ||||||||
| <30 | 354 | 83 | Ref | ||||||||
| 30-50 | 392 | 120 | 1.06 | 0.86-1331 | .59 | 1.12 | 0.84-1.49 | .40 | 1.27 | 0.88-1.83 | .194 |
| >50 | 104 | 43 | 1.55 | 1.15-2.10 | .004 | 1.61 | 1.08-2.40 | .02 | 2.14 | 1.31-3.50 | .002 |
| Donor type | |||||||||||
| MMUD | 195 | 46 | Ref | ||||||||
| Haplo | 660 | 200 | 1.17 | 0.91-1.51 | .21 | 1.21 | 0.92-1.59 | .16 | 1.20 | 0.83-1.72 | .33 |
| Disease | .01 | .02 | .05 | ||||||||
| ALL | 113 | 37 | Ref | ||||||||
| AML/MDS | 504 | 140 | 0.70 | 0.52-0.94 | .01 | 0.51 | 0.32-0.82 | .006 | 0.65 | 0.37-1.15 | .14 |
| Other | 238 | 69 | 0.71 | 0.51-0.98 | .04 | 0.54 | 0.33-0.90 | .01 | 1.00 | 0.56-1.79 | .99 |
| HCT-CI | .001 | .03 | .03 | ||||||||
| 0 | 162 | 38 | Ref | ||||||||
| 1-2 | 240 | 55 | 1.01 | 0.74-1.39 | .93 | 0.86 | 0.58-1.27 | .45 | 0.70 | 0.41-1.21 | .20 |
| ≥3 | 453 | 153 | 1.46 | 1.11-1.94 | .008 | 1.29 | 0.92-1.81 | .13 | 1.27 | 0.82-1.96 | .28 |
| D/R CMV | .95 | .06 | .33 | ||||||||
| Neg/Neg | 117 | 27 | Ref | ||||||||
| Neg/Pos | 268 | 82 | 1.40 | 1.00-1.95 | .04 | 1.66 | 1.10-2.49 | .01 | 1.63 | 0.95-2.78 | .07 |
| Pos/Neg | 77 | 18 | 0.91 | 0.58-1.44 | .69 | 1.14 | 0.67-1.96 | .62 | 1.31 | 0.67-2.57 | .43 |
| Pos/Pos | 393 | 119 | 1.17 | 0.85-1.62 | .33 | 1.22 | 0.82-1.83 | .32 | 1.30 | 0.78-2.18 | .31 |
| D/R sex mismatch | |||||||||||
| F-to-M | 179 | 52 | Ref | ||||||||
| Not F-to-M | 676 | 194 | 0.97 | 0.77-1.24 | .83 | 0.97 | 0.66-1.43 | .88 | 1.04 | 0.68-1.58 | .87 |
| Relapse† | |||||||||||
| Donor age, spline∗ | .79 | .69 | .59 | ||||||||
| Donor age, y∗ | .59 | .78 | .52 | ||||||||
| <30 | 354 | 101 | Ref | ||||||||
| 30-50 | 392 | 96 | 0.87 | 0.66-1.15 | .33 | 0.98 | 0.66-1.46 | .93 | 0.82 | 0.55-1.22 | .33 |
| ≥50 | 104 | 26 | 1.00 | 0.64-1.55 | .98 | 1.17 | 0.70-1.96 | .54 | 0.73 | 0.36-1.50 | .39 |
| Donor type | |||||||||||
| MMUD | 195 | 43 | Ref | ||||||||
| Haplo | 660 | 180 | 1.07 | 0.76-1.51 | .68 | 1.12 | 0.78-1.60 | .55 | 0.93 | 0.62-1.37 | .70 |
| DRI | <.001 | <.001 | <.001 | ||||||||
| Low | 71 | 13 | Ref | ||||||||
| Intermediate | 479 | 91 | 1.32 | 0.73-2.41 | .36 | 1.01 | 0.52-1.95 | .97 | 1.23 | 0.60-2.51 | .57 |
| High | 244 | 92 | 3.20 | 1.76-5.82 | <.001 | 3.01 | 1.57-5.79 | .001 | 2.73 | 1.33-5.60 | .006 |
| Very high | 42 | 24 | 8.10 | 4.05-16.20 | <.001 | 8.01 | 3.37-19.04 | <.001 | 12.38 | 4.89-31.34 | <.001 |
| Missing | 19 | 3 | 1.68 | 0.47-6.00 | .42 | 0.93 | 0.22-3.98 | .92 | 1.62 | 0.43-6.11 | .47 |
| D/R CMV | .28 | .03 | .83 | ||||||||
| Neg/Neg | 117 | 28 | Ref | ||||||||
| Neg/Pos | 268 | 81 | 1.25 | 0.81-1.93 | .32 | 1.12 | 0.66-1.89 | .67 | 1.00 | 0.56-1.78 | .96 |
| Pos/Neg | 77 | 17 | 0.95 | 0.52-1.76 | .87 | 0.49 | 0.24-1.01 | .054 | 0.67 | 0.28-1.63 | .38 |
| Pos/Pos | 393 | 97 | 0.93 | 0.61-1.42 | .72 | 0.72 | 0.43-1.20 | .20 | 0.97 | 0.56-1.68 | .91 |
| D/R sex mismatch | |||||||||||
| F-to-M | 179 | 45 | Ref | ||||||||
| Not F-to-M | 676 | 178 | 1.01 | 0.72-1.40 | .96 | 0.86 | 0.57-1.29 | .46 | 0.77 | 0.49-1.20 | .24 |
| Time to HCT, mo | .16 | .08 | .31 | ||||||||
| <6 | 225 | 48 | Ref | ||||||||
| 6-12 | 227 | 65 | 1.43 | 0.98-2.09 | .06 | 1.75 | 1.08-2.85 | .02 | 1.56 | 0.88-2.76 | .12 |
| >12 | 403 | 110 | 1.31 | 0.93-1.86 | .12 | 1.41 | 0.89-2.25 | .14 | 1.42 | 0.82-2.46 | .21 |
| aGVHD grade 3-4: AML/MDS | |||||||||||
| Donor age, spline∗ | .001 | .005 | .05 | ||||||||
| Donor age, y∗ | .0003 | .017 | .009 | ||||||||
| <30 | 197 | 10 | Ref | ||||||||
| 30-50 | 243 | 39 | 3.52 | 1.75-7.09 | <.001 | 2.05 | 0.93-4.52 | .07 | 2.70 | 1.22-5.96 | .01 |
| >50 | 61 | 11 | 5.36 | 2.20-13.04 | <.001 | 4.85 | 1.60-14.71 | .005 | 4.83 | 1.65-14.20 | .004 |
| Donor type | |||||||||||
| MMUD | 119 | 18 | Ref | ||||||||
| Haplo | 385 | 42 | 0.45 | 0.25-0.81 | .007 | 0.41 | 0.21-0.80 | .009 | 0.42 | 0.21-0.84 | .015 |
| D/R CMV | .22 | .003 | .16 | ||||||||
| Neg/Neg | 53 | 3 | Ref | ||||||||
| Neg/Pos | 175 | 20 | 2.86 | 0.84-9.70 | .09 | 12.18 | 3.06-48.51 | <.001 | 3.83 | 0.87-16.91 | .07 |
| Pos/Neg | 44 | 5 | 1.39 | 0.33-5.86 | .65 | 4.09 | 0.74-22.62 | .10 | 1.40 | 0.23-8.46 | .71 |
| Pos/Pos | 232 | 32 | 2.85 | 0.85-9.53 | .08 | 9.05 | 2.11-38.87 | .003 | 2.53 | 0.56-11.49 | .22 |
| HCT year | |||||||||||
| <2019 | 217 | 17 | Ref | ||||||||
| ≥2019 | 287 | 43 | 2.11 | 1.19-3.74 | .01 | 1.53 | 0.71-3.30 | .28 | 2.58 | 1.11-5.99 | .02 |
| aGVHD grade 3-4: not AML/MDS | |||||||||||
| Donor age, spline∗ | .66 | .77 | .73 | ||||||||
| Donor age, y∗ | .34 | .82 | .64 | ||||||||
| <30 | 157 | 18 | Ref | ||||||||
| 30-50 | 149 | 24 | 1.53 | 0.83-2.84 | .17 | 1.12 | 0.53-2.37 | .77 | 1.10 | 0.53-2.27 | .79 |
| >50 | 43 | 7 | 1.78 | 0.72-4.43 | .21 | 1.61 | 0.36-7.13 | .53 | 0.41 | 0.05-3.16 | .39 |
| Donor type | |||||||||||
| MMUD | 76 | 12 | Ref | ||||||||
| Haplo | 275 | 38 | 0.86 | 0.44-1.68 | .66 | 0.78 | 0.34-1.83 | .57 | 1.09 | 0.50-2.36 | .82 |
| D/R CMV | .16 | .04 | .22 | ||||||||
| Neg/Neg | 64 | 4 | Ref | ||||||||
| Neg/Pos | 93 | 16 | 3.61 | 1.19-10.98 | .02 | 6.84 | 1.58-29.64 | .01 | 3.36 | 1.07-10.57 | .03 |
| Pos/Neg | 33 | 6 | 2.65 | 0.74-9.53 | .13 | 6.82 | 1.33-34.89 | .02 | 2.72 | .67-11.02 | .16 |
| Pos/Pos | 161 | 24 | 2.88 | 0.99-8.39 | .052 | 4.14 | 0.86-19.87 | .07 | 2.20 | 0.70-6.93 | .17 |
| HCT year | |||||||||||
| <2019 | 176 | 27 | Ref | ||||||||
| ≥2019 | 175 | 23 | 1.10 | 0.63-1.94 | .73 | 1.78 | 0.80-3.93 | .15 | 1.25 | 0.59-2.63 | .56 |
| cGVHD‡ | |||||||||||
| Patient age, y | |||||||||||
| <50 | 387 | 56 | Ref | ||||||||
| ≥50 | 468 | 65 | 1.34 | 0.91-1.98 | .14 | 1.29 | 0.79-2.10 | .31 | 1.28 | 0.80-2.05 | .30 |
| Donor age, spline∗ | .07 | .06 | .33 | ||||||||
| Donor age, y∗ | .005 | .009 | .04 | ||||||||
| <30 | 354 | 39 | Ref | ||||||||
| 30-50 | 392 | 62 | 1.64 | 1.09-2.47 | .01 | 1.86 | 1.12-3.09 | .01 | 1.76 | 1.09-2.86 | .02 |
| >50 | 104 | 19 | 2.43 | 1.37-4.29 | .002 | 3.00 | 1.40-6.42 | .005 | 2.00 | 0.95-4.21 | .06 |
| Donor type | |||||||||||
| MMUD | 195 | 29 | Ref | ||||||||
| Haplo | 660 | 92 | 0.82 | 0.53-1.29 | .39 | 0.97 | 0.61-1.53 | .88 | 0.91 | 0.57-1.47 | .70 |
| D/R CMV | .24 | .72 | .71 | ||||||||
| Neg/Neg | 117 | 18 | Ref | ||||||||
| Neg/Pos | 268 | 29 | 0.79 | 0.43-1.44 | .43 | 0.86 | 0.43-1.73 | .67 | 0.81 | 0.41-1.60 | .54 |
| Pos/Neg | 77 | 9 | 0.63 | 0.28-1.40 | .25 | 0.57 | 0.22-1.49 | .25 | 0.87 | 0.36-2.08 | .75 |
| Pos/Pos | 393 | 65 | 1.16 | 0.67-2.00 | .59 | 1.03 | 0.53-1.99 | .93 | 1.11 | 0.59-2.07 | .75 |
| Graft type | |||||||||||
| Bone marrow | 537 | 64 | Ref | ||||||||
| Peripheral blood | 318 | 57 | 1.90 | 1.30-2.79 | .001 | 1.61 | 0.99-2.62 | .054 | 1.90 | 1.18-3.06 | .008 |
| D/R sex mismatch | |||||||||||
| F-to-M | 179 | 27 | Ref | ||||||||
| Not F-to-M | 676 | 94 | 1.17 | 0.76-1.82 | .47 | 1.60 | 0.91-2.81 | .10 | 1.71 | 0.93-3.14 | .08 |
| . | N . | Events . | Cox PH (entire cohort) . | IPTW analysis (entire cohort) . | Cox PH (matched-pair cohort) . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HR . | 95% CI . | P value . | HR . | 95% CI . | P value . | HR . | 95% CI . | P value . | |||
| OS | |||||||||||
| Recipient age, y | |||||||||||
| <50 | 387 | 160 | Ref | ||||||||
| ≥50 | 468 | 265 | 1.70 | 1.39-2.09 | <.001 | 1.82 | 1.36-2.43 | <.001 | 1.85 | 1.38-2.47 | <.001 |
| Donor age, spline∗ | .05 | .049 | .05 | ||||||||
| Donor age, y∗ | .03 | .08 | .03 | ||||||||
| <30 | 354 | 163 | Ref | ||||||||
| 30-50 | 392 | 196 | 1.04 | 0.84-1.29 | .72 | 1.09 | 0.82-1.45 | .53 | 1.02 | 0.77-1.36 | .87 |
| >50 | 104 | 66 | 1.46 | 1.09-1.97 | .01 | 1.52 | 1.05-2.22 | .02 | 1.68 | 1.13-2.50 | .01 |
| Donor type | |||||||||||
| MMUD | 195 | 78 | Ref | ||||||||
| Haplo | 660 | 347 | 1.20 | 0.93-1.54 | .16 | 1.22 | 0.93-1.60 | .15 | 1.15 | 0.87-1.52 | .32 |
| Disease | .07 | ||||||||||
| ALL | 113 | 63 | Ref | ||||||||
| AML/MDS | 504 | 254 | - | - | - | 0.64 | 0.39-1.06 | .08 | - | - | - |
| Other | 238 | 108 | - | - | - | 0.88 | 0.51-1.51 | .64 | - | - | - |
| DRI | <.001 | <.001 | <.001 | ||||||||
| Low | 71 | 24 | Ref | ||||||||
| Intermediate | 479 | 201 | 1.23 | 0.80-1.90 | .35 | 1.48 | 0.80-2.72 | .21 | 1.19 | 0.70-2.00 | .51 |
| High | 244 | 154 | 2.13 | 1.37-3.31 | .001 | 2.77 | 1.46-5.26 | .002 | 1.88 | 1.09-3.24 | .02 |
| Very high | 42 | 36 | 4.20 | 2.48-7.11 | <.001 | 4.91 | 2.41-10.01 | <.001 | 4.84 | 2.39-9.78 | <.001 |
| Missing | 19 | 10 | 2.05 | 0.97-4.31 | .06 | 2.46 | 1.04-5.85 | .04 | 2.08 | 0.94-4.61 | .07 |
| HCT-CI | .002 | .04 | .11 | ||||||||
| 0 | 162 | 63 | Ref | ||||||||
| 1-2 | 240 | 107 | 1.00 | 0.73-1.37 | .99 | 0.89 | 0.60-1.30 | .54 | 0.87 | 0.58-1.31 | .50 |
| ≥3 | 453 | 255 | 1.42 | 1.07-1.88 | .01 | 1.28 | 0.91-1.81 | .16 | 1.22 | 0.86-1.73 | .26 |
| D/R CMV | .14 | .06 | .53 | ||||||||
| Neg/Neg | 117 | 48 | Ref | ||||||||
| Neg/Pos | 268 | 148 | 1.33 | 0.96-1.85 | .09 | 1.55 | 1.03-2.34 | .03 | 1.30 | 0.86-1.97 | .21 |
| Pos/Neg | 77 | 31 | 0.92 | 0.58-1.46 | .73 | 1.11 | 0.65-1.89 | .69 | 1.00 | 0.57-1.75 | .99 |
| Pos/Pos | 393 | 198 | 1.12 | 0.81-1.55 | .48 | 1.11 | 0.74-1.66 | .61 | 1.13 | 0.76-1.69 | .55 |
| D/R sex mismatch | |||||||||||
| F-to-M | 676 | 337 | Ref | ||||||||
| Not F-to-M | 179 | 88 | 0.94 | 0.74-1.19 | .61 | 0.92 | 0.69-1.24 | .58 | 0.86 | 0.62-1.18 | .35 |
| NRM† | |||||||||||
| Recipient age, y | |||||||||||
| <50 | 387 | 77 | Ref | ||||||||
| ≥50 | 468 | 169 | 1.78 | 1.44-2.20 | <.001 | 1.77 | 1.33-2.38 | <.001 | 2.25 | 1.52-3.33 | <.001 |
| Donor age, spline∗ | .036 | .02 | .06 | ||||||||
| Donor age, y∗ | .02 | .06 | .009 | ||||||||
| <30 | 354 | 83 | Ref | ||||||||
| 30-50 | 392 | 120 | 1.06 | 0.86-1331 | .59 | 1.12 | 0.84-1.49 | .40 | 1.27 | 0.88-1.83 | .194 |
| >50 | 104 | 43 | 1.55 | 1.15-2.10 | .004 | 1.61 | 1.08-2.40 | .02 | 2.14 | 1.31-3.50 | .002 |
| Donor type | |||||||||||
| MMUD | 195 | 46 | Ref | ||||||||
| Haplo | 660 | 200 | 1.17 | 0.91-1.51 | .21 | 1.21 | 0.92-1.59 | .16 | 1.20 | 0.83-1.72 | .33 |
| Disease | .01 | .02 | .05 | ||||||||
| ALL | 113 | 37 | Ref | ||||||||
| AML/MDS | 504 | 140 | 0.70 | 0.52-0.94 | .01 | 0.51 | 0.32-0.82 | .006 | 0.65 | 0.37-1.15 | .14 |
| Other | 238 | 69 | 0.71 | 0.51-0.98 | .04 | 0.54 | 0.33-0.90 | .01 | 1.00 | 0.56-1.79 | .99 |
| HCT-CI | .001 | .03 | .03 | ||||||||
| 0 | 162 | 38 | Ref | ||||||||
| 1-2 | 240 | 55 | 1.01 | 0.74-1.39 | .93 | 0.86 | 0.58-1.27 | .45 | 0.70 | 0.41-1.21 | .20 |
| ≥3 | 453 | 153 | 1.46 | 1.11-1.94 | .008 | 1.29 | 0.92-1.81 | .13 | 1.27 | 0.82-1.96 | .28 |
| D/R CMV | .95 | .06 | .33 | ||||||||
| Neg/Neg | 117 | 27 | Ref | ||||||||
| Neg/Pos | 268 | 82 | 1.40 | 1.00-1.95 | .04 | 1.66 | 1.10-2.49 | .01 | 1.63 | 0.95-2.78 | .07 |
| Pos/Neg | 77 | 18 | 0.91 | 0.58-1.44 | .69 | 1.14 | 0.67-1.96 | .62 | 1.31 | 0.67-2.57 | .43 |
| Pos/Pos | 393 | 119 | 1.17 | 0.85-1.62 | .33 | 1.22 | 0.82-1.83 | .32 | 1.30 | 0.78-2.18 | .31 |
| D/R sex mismatch | |||||||||||
| F-to-M | 179 | 52 | Ref | ||||||||
| Not F-to-M | 676 | 194 | 0.97 | 0.77-1.24 | .83 | 0.97 | 0.66-1.43 | .88 | 1.04 | 0.68-1.58 | .87 |
| Relapse† | |||||||||||
| Donor age, spline∗ | .79 | .69 | .59 | ||||||||
| Donor age, y∗ | .59 | .78 | .52 | ||||||||
| <30 | 354 | 101 | Ref | ||||||||
| 30-50 | 392 | 96 | 0.87 | 0.66-1.15 | .33 | 0.98 | 0.66-1.46 | .93 | 0.82 | 0.55-1.22 | .33 |
| ≥50 | 104 | 26 | 1.00 | 0.64-1.55 | .98 | 1.17 | 0.70-1.96 | .54 | 0.73 | 0.36-1.50 | .39 |
| Donor type | |||||||||||
| MMUD | 195 | 43 | Ref | ||||||||
| Haplo | 660 | 180 | 1.07 | 0.76-1.51 | .68 | 1.12 | 0.78-1.60 | .55 | 0.93 | 0.62-1.37 | .70 |
| DRI | <.001 | <.001 | <.001 | ||||||||
| Low | 71 | 13 | Ref | ||||||||
| Intermediate | 479 | 91 | 1.32 | 0.73-2.41 | .36 | 1.01 | 0.52-1.95 | .97 | 1.23 | 0.60-2.51 | .57 |
| High | 244 | 92 | 3.20 | 1.76-5.82 | <.001 | 3.01 | 1.57-5.79 | .001 | 2.73 | 1.33-5.60 | .006 |
| Very high | 42 | 24 | 8.10 | 4.05-16.20 | <.001 | 8.01 | 3.37-19.04 | <.001 | 12.38 | 4.89-31.34 | <.001 |
| Missing | 19 | 3 | 1.68 | 0.47-6.00 | .42 | 0.93 | 0.22-3.98 | .92 | 1.62 | 0.43-6.11 | .47 |
| D/R CMV | .28 | .03 | .83 | ||||||||
| Neg/Neg | 117 | 28 | Ref | ||||||||
| Neg/Pos | 268 | 81 | 1.25 | 0.81-1.93 | .32 | 1.12 | 0.66-1.89 | .67 | 1.00 | 0.56-1.78 | .96 |
| Pos/Neg | 77 | 17 | 0.95 | 0.52-1.76 | .87 | 0.49 | 0.24-1.01 | .054 | 0.67 | 0.28-1.63 | .38 |
| Pos/Pos | 393 | 97 | 0.93 | 0.61-1.42 | .72 | 0.72 | 0.43-1.20 | .20 | 0.97 | 0.56-1.68 | .91 |
| D/R sex mismatch | |||||||||||
| F-to-M | 179 | 45 | Ref | ||||||||
| Not F-to-M | 676 | 178 | 1.01 | 0.72-1.40 | .96 | 0.86 | 0.57-1.29 | .46 | 0.77 | 0.49-1.20 | .24 |
| Time to HCT, mo | .16 | .08 | .31 | ||||||||
| <6 | 225 | 48 | Ref | ||||||||
| 6-12 | 227 | 65 | 1.43 | 0.98-2.09 | .06 | 1.75 | 1.08-2.85 | .02 | 1.56 | 0.88-2.76 | .12 |
| >12 | 403 | 110 | 1.31 | 0.93-1.86 | .12 | 1.41 | 0.89-2.25 | .14 | 1.42 | 0.82-2.46 | .21 |
| aGVHD grade 3-4: AML/MDS | |||||||||||
| Donor age, spline∗ | .001 | .005 | .05 | ||||||||
| Donor age, y∗ | .0003 | .017 | .009 | ||||||||
| <30 | 197 | 10 | Ref | ||||||||
| 30-50 | 243 | 39 | 3.52 | 1.75-7.09 | <.001 | 2.05 | 0.93-4.52 | .07 | 2.70 | 1.22-5.96 | .01 |
| >50 | 61 | 11 | 5.36 | 2.20-13.04 | <.001 | 4.85 | 1.60-14.71 | .005 | 4.83 | 1.65-14.20 | .004 |
| Donor type | |||||||||||
| MMUD | 119 | 18 | Ref | ||||||||
| Haplo | 385 | 42 | 0.45 | 0.25-0.81 | .007 | 0.41 | 0.21-0.80 | .009 | 0.42 | 0.21-0.84 | .015 |
| D/R CMV | .22 | .003 | .16 | ||||||||
| Neg/Neg | 53 | 3 | Ref | ||||||||
| Neg/Pos | 175 | 20 | 2.86 | 0.84-9.70 | .09 | 12.18 | 3.06-48.51 | <.001 | 3.83 | 0.87-16.91 | .07 |
| Pos/Neg | 44 | 5 | 1.39 | 0.33-5.86 | .65 | 4.09 | 0.74-22.62 | .10 | 1.40 | 0.23-8.46 | .71 |
| Pos/Pos | 232 | 32 | 2.85 | 0.85-9.53 | .08 | 9.05 | 2.11-38.87 | .003 | 2.53 | 0.56-11.49 | .22 |
| HCT year | |||||||||||
| <2019 | 217 | 17 | Ref | ||||||||
| ≥2019 | 287 | 43 | 2.11 | 1.19-3.74 | .01 | 1.53 | 0.71-3.30 | .28 | 2.58 | 1.11-5.99 | .02 |
| aGVHD grade 3-4: not AML/MDS | |||||||||||
| Donor age, spline∗ | .66 | .77 | .73 | ||||||||
| Donor age, y∗ | .34 | .82 | .64 | ||||||||
| <30 | 157 | 18 | Ref | ||||||||
| 30-50 | 149 | 24 | 1.53 | 0.83-2.84 | .17 | 1.12 | 0.53-2.37 | .77 | 1.10 | 0.53-2.27 | .79 |
| >50 | 43 | 7 | 1.78 | 0.72-4.43 | .21 | 1.61 | 0.36-7.13 | .53 | 0.41 | 0.05-3.16 | .39 |
| Donor type | |||||||||||
| MMUD | 76 | 12 | Ref | ||||||||
| Haplo | 275 | 38 | 0.86 | 0.44-1.68 | .66 | 0.78 | 0.34-1.83 | .57 | 1.09 | 0.50-2.36 | .82 |
| D/R CMV | .16 | .04 | .22 | ||||||||
| Neg/Neg | 64 | 4 | Ref | ||||||||
| Neg/Pos | 93 | 16 | 3.61 | 1.19-10.98 | .02 | 6.84 | 1.58-29.64 | .01 | 3.36 | 1.07-10.57 | .03 |
| Pos/Neg | 33 | 6 | 2.65 | 0.74-9.53 | .13 | 6.82 | 1.33-34.89 | .02 | 2.72 | .67-11.02 | .16 |
| Pos/Pos | 161 | 24 | 2.88 | 0.99-8.39 | .052 | 4.14 | 0.86-19.87 | .07 | 2.20 | 0.70-6.93 | .17 |
| HCT year | |||||||||||
| <2019 | 176 | 27 | Ref | ||||||||
| ≥2019 | 175 | 23 | 1.10 | 0.63-1.94 | .73 | 1.78 | 0.80-3.93 | .15 | 1.25 | 0.59-2.63 | .56 |
| cGVHD‡ | |||||||||||
| Patient age, y | |||||||||||
| <50 | 387 | 56 | Ref | ||||||||
| ≥50 | 468 | 65 | 1.34 | 0.91-1.98 | .14 | 1.29 | 0.79-2.10 | .31 | 1.28 | 0.80-2.05 | .30 |
| Donor age, spline∗ | .07 | .06 | .33 | ||||||||
| Donor age, y∗ | .005 | .009 | .04 | ||||||||
| <30 | 354 | 39 | Ref | ||||||||
| 30-50 | 392 | 62 | 1.64 | 1.09-2.47 | .01 | 1.86 | 1.12-3.09 | .01 | 1.76 | 1.09-2.86 | .02 |
| >50 | 104 | 19 | 2.43 | 1.37-4.29 | .002 | 3.00 | 1.40-6.42 | .005 | 2.00 | 0.95-4.21 | .06 |
| Donor type | |||||||||||
| MMUD | 195 | 29 | Ref | ||||||||
| Haplo | 660 | 92 | 0.82 | 0.53-1.29 | .39 | 0.97 | 0.61-1.53 | .88 | 0.91 | 0.57-1.47 | .70 |
| D/R CMV | .24 | .72 | .71 | ||||||||
| Neg/Neg | 117 | 18 | Ref | ||||||||
| Neg/Pos | 268 | 29 | 0.79 | 0.43-1.44 | .43 | 0.86 | 0.43-1.73 | .67 | 0.81 | 0.41-1.60 | .54 |
| Pos/Neg | 77 | 9 | 0.63 | 0.28-1.40 | .25 | 0.57 | 0.22-1.49 | .25 | 0.87 | 0.36-2.08 | .75 |
| Pos/Pos | 393 | 65 | 1.16 | 0.67-2.00 | .59 | 1.03 | 0.53-1.99 | .93 | 1.11 | 0.59-2.07 | .75 |
| Graft type | |||||||||||
| Bone marrow | 537 | 64 | Ref | ||||||||
| Peripheral blood | 318 | 57 | 1.90 | 1.30-2.79 | .001 | 1.61 | 0.99-2.62 | .054 | 1.90 | 1.18-3.06 | .008 |
| D/R sex mismatch | |||||||||||
| F-to-M | 179 | 27 | Ref | ||||||||
| Not F-to-M | 676 | 94 | 1.17 | 0.76-1.82 | .47 | 1.60 | 0.91-2.81 | .10 | 1.71 | 0.93-3.14 | .08 |
F, female; M, male; Ref, reference.
Two Cox PH models were built with identical covariates except for donor age: 1 with categorical groups (donor age of <30, 30-50, and >50 years), and another using natural cubic spline (4 knots) for nonlinear effects. Both results are shown in 1 table for brevity.
Relapse model was stratified by conditioning intensity (violation of PH assumptions).
cGVHD model was stratified by disease type (violation of PH assumptions).
Effect of donor age on OS. The nonlinear effect of donor age on OS is visualized using a natural spline from a Cox PH multivariate model (adjusted covariates shown in Table 2), with HRs plotted relative to a donor age of 34 years (median donor age).
Effect of donor age on OS. The nonlinear effect of donor age on OS is visualized using a natural spline from a Cox PH multivariate model (adjusted covariates shown in Table 2), with HRs plotted relative to a donor age of 34 years (median donor age).
Assessment of interaction effects did not reveal strong evidence of effect modification by donor type for donor age or other covariates. Specifically, in categorical models, interaction terms for donor age (>50 years) and haploidentical donor, and donor age (30-50 years) and haploidentical donor, yielded P values of .82 and .59, respectively. The LRT for the overall interaction between categorical donor age and donor type resulted in P value of .78. Similarly, in spline models, the LRT for the interaction between the natural spline of donor age and donor type showed a P value of .18. Spline curves of donor age and overall mortality hazard by donor type (MMUD vs haploidentical) from the multivariate model are shown in supplemental Figure 4.
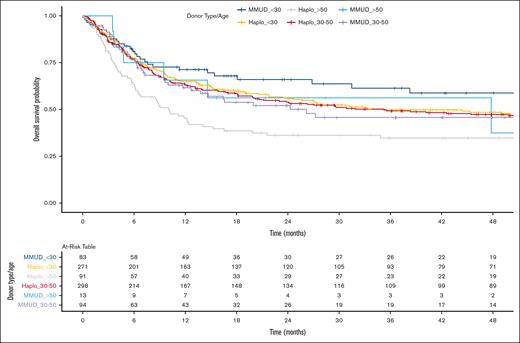
We conducted an exploratory analysis to directly compare outcomes between haploidentical and MMUD groups, further categorized by donor age (<30 years, 30-50 years, and >50 years; Figure 3. In a Cox PH model, without adjustment for multiple comparisons, the HR for haploidentical donors aged >50 years vs MMUD donors aged <30 years (reference) was 1.91 (95% CI, 1.21-3.01; P = .005). HRs for other groups compared with the reference ranged from 1.20 to 1.30 (P > .05). These findings were further supported by the IPTW and the matched-pair analyses (supplemental Table 3).
Kaplan-Meier estimate of OS comparing Haplo donor and MMUD groups, further categorized by donor age (<30 years, 30-50 years, and >50 years).
Kaplan-Meier estimate of OS comparing Haplo donor and MMUD groups, further categorized by donor age (<30 years, 30-50 years, and >50 years).
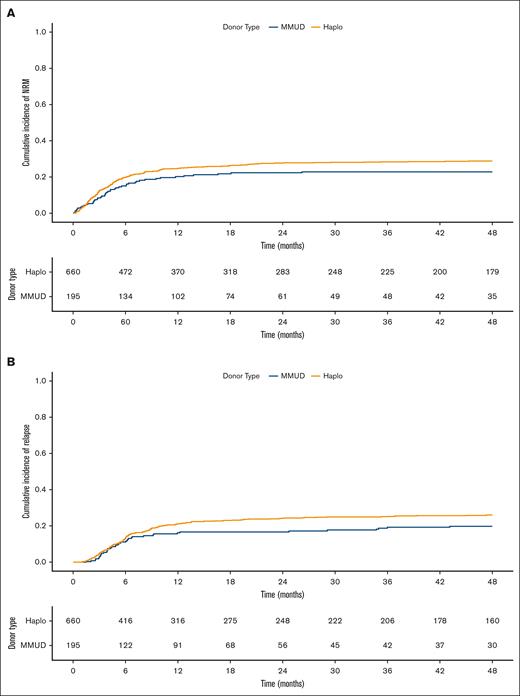
NRM and relapse
The cumulative incidence of NRM and relapse are shown in Figure 4. HRs for donors aged >50 years compared with those aged <30 years (reference), ranged from 1.55 to 2.14 in categorical models, and spline models indicating a negative impact of donor age and higher NRM with global P values of .02 to .06. The HR for donor type, haploidentical vs MMUD (reference) was 1.17 (95% CI, 0.91-1.51; P = .21) in the Cox PH model, with similar HRs in other models (Table 2).
NRM and relapse by donor type. Cumulative incidence of NRM (A) and relapse (B) comparing Haplo donor (yellow line) and MMUD (blue line) groups.
NRM and relapse by donor type. Cumulative incidence of NRM (A) and relapse (B) comparing Haplo donor (yellow line) and MMUD (blue line) groups.
For relapse, neither donor type nor donor age showed a notable association when donor age was modeled as a categorical variable or as spline (global P = .59-.79; Table 2). A nonlinear relationship between donor age and the hazard of both NRM and relapse was noted, as estimated by separate Cox PH MVA using natural splines (supplemental Figure 5). Although NRM hazard showed a generally increasing trend with donor age, relapse hazard, exhibited a more complex nonmonotonic relationship with donor age. Additionally, to exclude disease heterogeneity as a confounder, we performed a subgroup analysis restricted to patients with AML/MDS and demonstrated similar findings for the risks of NRM and relapse (data not shown).
aGVHD and cGVHD
Analyses of grade 3 to 4 aGVHD revealed an interaction between donor type and disease. Patients with older donors (aged >50 years) had an approximate fivefold higher risk of grade 3 to 4 aGVHD than those with younger (aged <30 years) donors. Haploidentical donors were associated with ∼55% to 60% lower risk than the MMUD group in AML/MDS, but not in other diseases (Table 2). In contrast, in MVA of cGVHD, donor type did not show a notable influence, whereas older donor age (aged >50 years) was associated with approximately twofold to threefold higher hazard (Table 2).
Other outcomes
Of 195 patients in the MMUD group, 2 (1%) did not achieve engraftment and 7 (∼4%) died before engraftment. In the haploidentical donor group, 16 of 660 (∼2%) did not achieve engraftment and 19 of 660 (∼3%) died before engraftment. The most common cause of death in both donor groups was the relapse of the underlying malignancy accounting for 31 of 78 (∼40%) in the MMUD group and 144 of 345 (42%) in the haploidentical group. This was followed by infections, which accounted for 19 of 78 (∼24%) in MMUD and 88 of 345 (∼26%) in the haploidentical group (supplemental Table 4).
Validation analysis using external CIBMTR cohort
Baseline characteristics of the entire cohort used for sensitivity analysis (haploidentical MDACC, n = 236; MMUD-CIBMTR, n = 458) and 1:1 matched-pair cohort (haploidentical MDACC = 107, MMUD-CIBMTR = 107) are shown in supplemental Table 5. The Cox PH MVA of overall mortality revealed an association with donor age, modeled as spline (P = .017). The HR for haploidentical was 1.26 (95% CI, 0.83-1.95; P = .29) compared with the MMUD group (supplemental Table 6). Also, the nonlinear components of the donor age spline indicated that the nonlinear specification better aligns with the data than a linear term alone.
Discussion
Our analysis comparing the outcomes of the 2 most commonly pursued mismatched donors, haploidentical and MMUDs, in the era of PTCy, suggests that donor age may be a more influential factor than donor type as the main effect. However, the role of donor type appears more nuanced, potentially influenced by donor age, with effects becoming more evident within older donor cohorts. Although statistical modeling of interactions did not reveal strong evidence of interaction, our exploratory analyses, such as the comparison of specific donor type and donor age groups and spline model stratified by donor type, suggest a more complex relationship, suggesting potential combined effects of donor age and donor type.
Considering that the unrelated donor pool generally consists of younger individuals and that haploidentical donors can encompass a broader age range, it might be more clinically impactful to highlight the potential drawbacks of older haploidentical donors rather than reiterating the selection of younger MMUDs, a practice that is steadily gaining traction. In patients for whom haploidentical donors are older, expanding the search to second-degree relatives could identify more suitable donors. Several studies have reported the safety and feasibility of using second-degree haploidentical donors, including grandchildren, nephews, and nieces who are more likely to be younger.20-22
Our study has limitations that warrant consideration. Firstly, the relatively smaller size of the MMUD group precluded a comprehensive investigation of the impact of independent HLA factors such as antigen vs allele matching, peptide-binding motif matching vs mismatching, and class 1 vs class 2 mismatches within this group, which may affect the outcomes.23-26 However, the specific impact of HLA mismatches in MMUDs with PTCy prophylaxis remains undefined. Recent CIBMTR data suggest that PTCy may mitigate HLA mismatch effects, resulting in MMUD outcomes comparable with those of matched unrelated donors.9 Similarly, a large European Group for Blood and Marrow Transplantation study could not conclusively determine whether any single locus mismatch is superior when PTCy prophylaxis is used.24 This evidence guided our focus on non-HLA factors, specifically donor age in both donor types, in our single-institution analysis. Similarly, the influence of individual HLA factors within the haploidentical cohort, which has been described to be associated with survival outcomes,27 was not assessed in this study. The importance of considering HLA factors in comparing donor types was highlighted in a previous study that evaluated the outcomes of MMUD (categorized by peptide-binding motif matching) vs haploidentical donors (categorized by B-leader and DRB1 matching). However, MMUD recipients in that study received calcineurin inhibitor–based GVHD prophylaxis.26 Therefore, future studies should investigate the impact of these 2 donor types with PTCy prophylaxis, stratified by their prognostic HLA factors.
Next, the binary categorization of race (White vs non-White) represents a simplification of complex racial and ethnic diversity. This approach was necessitated by sample size limitations within specific subgroups, especially in the MMUD cohort, which precluded a more detailed analysis. Transplant year was dichotomized at 2019, the median year in the MDACC cohort, which may have oversimplified temporal trends in supportive care. Similarly, as the dichotomization of patient age was arbitrary, future studies could explore continuous modeling to better capture patient age–related effects. Also, we lack data on steroid-refractory aGVHD or cGVHD, the National Institutes of Health-defined cGVHD, and therapy-requiring cGVHD. Additionally, the impact of donor relationship in the haploidentical group, which has been extensively studied previously,22,28-30 was not reexamined in our study. Our analysis incorporated key donor-related factors, including donor age, donor-recipient sex mismatch, and CMV serostatus, which appeared to influence outcomes, particularly NRM, especially in CMV-seropositive recipients. However, data limitations, notably the lack of information on letermovir prophylaxis, prevented further exploration and a detailed examination of its interactions with other donor-recipient factors. In addition, information about donor-specific anti-HLA antibodies was not available. Nevertheless, we generally avoid using donors against whom the recipient carries donor-specific anti-HLA antibodies, making it less likely to have introduced a major bias in this context. Lastly, the retrospective nature of our study introduces inherent biases associated with such study designs.
Our study possesses several key strengths. As a single-center study with standardized treatment protocols, such as infection prevention, GVHD prophylaxis, and management of posttransplant complications, it minimizes the variability in clinical practice. Although acknowledging potential baseline differences between donor groups, we used robust statistical methods to mitigate confounding factors. These included IPTW and matched-pair analyses in addition to the standard Cox PH models, all of which revealed consistent findings. Furthermore, we incorporated a data set from the CIBMTR for further analysis. This approach provides cross-validation of our findings and addresses some limitations of single-center data, such as limited donor diversity and small sample size, thereby enhancing the generalizability and robustness of our results.
Conclusions
Acknowledging the limitations of a single-center analysis, our study of PTCy-based HCT revealed a nuanced interplay between donor type (haploidentical vs MMUD) and donor age in influencing outcomes. Although donor age emerged as a more consistently influential predictor of OS, the role of donor type appears more complex, potentially modulated by donor age and becoming more pronounced in older donor cohorts. Further research using larger data sets is essential to fully elucidate the potential interplay between donor type and donor age, and to comprehensively assess the impact of HLA disparities within the haploidentical and MMUD groups on HCT outcomes.
Acknowledgments
A part of this study included a data set from the Center for International Blood and Marrow Transplant Research, which is supported primarily by the Public Health Service U24CA076518 from the National Cancer Institute; the National Heart, Lung, and Blood Institute; the National Institute of Allergy and Infectious Diseases; 75R60222C00011 from the Health Resources and Services Administration; N00014-21-1-2954 and N00014-23-1-2057 from the Office of Naval Research; the National Marrow Donor Program; and the Medical College of Wisconsin.
Authorship
Contribution: Y.M.A. abstracted the study design, interpreted the data, and wrote the manuscript; R.S.M. conceptualized and designed the study, performed the statistical analysis, interpreted data, helped with the manuscript writing, and had the final responsibility to submit for publication; J.R., G.R., P.S., P.K., U.P., B.O., K.R., R.E.C., and E.J.S. reviewed and interpreted the data, reviewed the manuscript, and provided critical feedback; Y.M.A., G.R., and R.S.M. had full access to the raw data; and all authors approved the manuscript.
Conflict-of-interest disclosure: K.R. and The University of Texas MD Anderson Cancer Center have an institutional financial conflict of interest with Takeda Pharmaceutical and Affimed GmbH. K.R. participates on the scientific advisory board for Avenge Bio, Virogin Biotech, Navan Technologies, Caribou Biosciences, Bit Bio Limited, Replay Holdings, oNKo-innate, the Alliance for Cancer Gene Therapy, Innate Pharma, and Shinobi Therapeutics; and is the scientific founder of Syena. The remaining authors declare no competing financial interests.
Correspondence: Rohtesh S. Mehta, MD Anderson Cancer Center, 1515 Holcombe Blvd, Unit 0423, Houston, TX 77030; email: rmehta1@mdanderson.org.
References
Author notes
The Center for International Blood and Marrow Transplant Research data used in this study are publicly available and accessible at https://cibmtr.org/CIBMTR/Resources/Publicly-Available-Datasets.
Original data may be available on request from the corresponding author, Rohtesh S. Mehta (rmehta1@mdanderson.org).
The full-text version of this article contains a data supplement.