Key Points
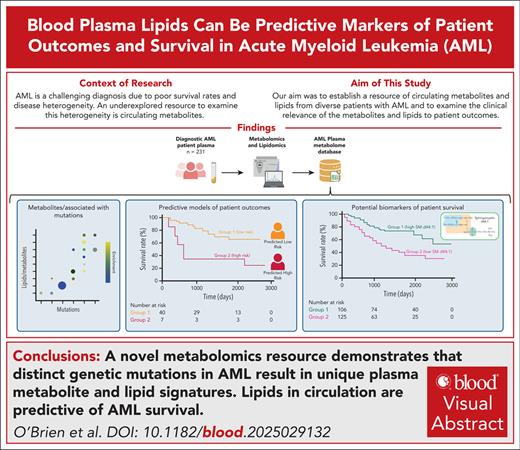
A novel metabolomics resource demonstrates that distinct genetic mutations in AML result in unique plasma metabolite and lipid signatures.
Lipids in circulation are predictive of AML survival.
Visual Abstract
Acute myeloid leukemia (AML) is characterized by a low 5-year survival rate. Despite having many clinical metrics to assess patient prognosis, there remain opportunities to improve risk stratification. We hypothesized that an underexplored resource to examine the prognosis of patients with AML is plasma metabolome. Circulating metabolites are influenced by patients’ clinical status and can serve as accessible cancer biomarkers. To establish a resource of circulating metabolites in genetically diverse patients with AML, we performed an unbiased metabolomic and lipidomic analysis of 231 diagnostic AML plasma samples before treatment with intensive chemotherapy. Intriguingly, circulating metabolites were highly associated with the mutation status within the AML cells. Furthermore, lipids were associated with refractory status. We established a machine learning algorithm trained on chemotherapy-refractory–associated lipids to predict patient survival. Cox regression and Kaplan-Meier analysis demonstrated that the high-risk lipid signature predicted overall survival in this patient cohort. Impressively, the top lipid in the high-risk lipid signature, sphingomyelin (d44:1), was sufficient to predict overall survival in both the original data set and an independent validation data set. Overall, this research underscores the potential of circulating metabolites to capture AML heterogeneity and lipids to be used as potential AML biomarkers.
Introduction
Despite recent advances in understanding the biological basis of acute myeloid leukemia (AML), outcomes for patients with this disease remain poor, partly due to limited therapeutic efficacy targeting subclonal heterogeneity.1 Despite recent therapeutic advances involving targeted therapies, intensive chemotherapy remains an important mainstay of the treatment of this disease. Although a multitude of metrics exist to risk stratify patients, further refinement of these strategies is still needed.2 Initial response to chemotherapy is a strong predictor of patient outcomes.3 Chemotherapy nonresponse (refractory AML) is defined as a lack of partial or complete remission after 2 courses of induction chemotherapy.4 However, by definition, patients with refractory AML are not recognized until after failure of induction therapy.5 The ability to predict refractory disease before the administration of chemotherapy could improve patient outcomes and quality of life by avoiding exposure to chemotherapy for patients unlikely to respond and who may be better candidates for other approved or experimental therapies.
To improve patient stratification, efforts are underway to examine the heterogeneity of patients with AML using omics techniques.6 A readily available and minimally invasive resource to examine the heterogeneity of patients with AML is the peripheral blood plasma metabolome. The components and origin of the circulating metabolome are diverse. Most metabolites are derived from tissue processes, with major contributions coming from the gut, muscles, and kidneys.7 Variability in plasma metabolites can be linked to diet,8 microbiome composition of the gut,9 lifestyle choices,10 and environmental factors.11 Time of day when samples are collected can be an additional confounding factor.12 Despite the many potential sources of variability, detailed analyses investigating the contributors to circulating metabolites in blood have demonstrated that clinical patient data are the best predictors of lipids, amino acids, and peptides, compared to other contributors to the circulating metabolome.13 Thus, surveying the circulating metabolome across heterogenous AML populations may reveal novel biomarkers in AML.
Research has sought to identify circulating metabolic biomarkers of multiple cancers including leukemia.6,14-21 However, a comprehensive survey of circulating lipids and metabolites has not yet been undertaken in AML. Therefore, we examined the predictive power of the circulating plasma lipidome in patients with AML at diagnosis before intensive chemotherapy. Using unbiased lipidomics and metabolomics, we captured metabolic heterogeneity associated with diverse disease parameters. Importantly, we established a lipid signature predictive of overall survival using a machine learning gradient boosted classifier (GBC) trained on lipids affiliated with refractory disease. The lipid most predictive of patient response, sphingomyelin (SM; d44:1), was independently sufficient to predict overall patient survival in the original data set as well as a new and distinct validation data set.
Study design
Plasma samples and clinical data
A total of 231 clinically annotated patient AML plasma samples, collected at diagnosis, were obtained from the Princess Margaret Biobank in accordance with the research ethics board of the University Health Network (protocol 20-503). A validation cohort of 67 plasma samples from patients with AML was obtained from the University of Colorado hematologic malignancies tissue bank. Details on plasma samples are provided in the supplemental Methods, available on the Blood website.
Metabolomics/lipidomics
Plasma samples were aliquoted on ice into microfuge tubes in 50-μL volumes. Samples were flash frozen in liquid nitrogen vapors, before storage at –80°C. Metabolomic and lipidomic analyses were performed by mass spectrometry as described previously.22 Detailed methods provided in the supplemental Methods.
Data analysis
Metabolites and metabolic pathways were analyzed using MetaboAnalystR (version 4.0),23 LipidR, LIPEA,24 and BioPAN.25 Machine learning models were developed using scikit-learn in python, and Cox regression analysis was performed using survminer and survival packages in R. Detailed methods are provided in the supplemental Methods.
Results and discussion
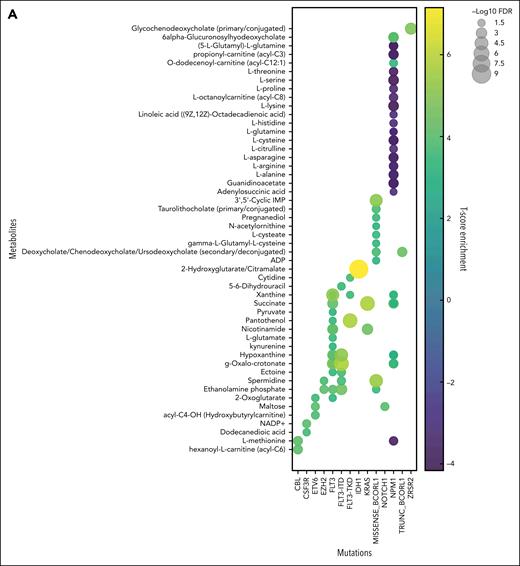
From 231 diagnosis patient samples (the Princess Margaret Hospital [PMH] data set), annotation of 17 diagnostically relevant clinical features (Table 1) as well as 66 cytogenetic and molecular abnormalities commonly observed in AML (supplemental Table 1) was performed. Metabolite levels from each plasma sample were interrogated by mass spectrometry, identifying 177 metabolites and 1988 lipids across all samples (supplemental Tables 2 and 3). Metabolites and metabolic pathways were further analyzed using MetaboAnalystR, LIPEA, or BioPAN. Of the 17 clinical features assessed, 5 had significant associations with specific metabolites (supplemental Figure 1A), and 11 had significant association with specific lipids (supplemental Figure 1B). Response to induction 7+3 therapy resulted in hundreds of significantly different lipids between responders and nonresponders (supplemental Table 4). Nonresponders were defined as patients who did not achieve a complete response after 2 cycles of induction chemotherapy, except for 2 patients who died <30 days after diagnosis who were also characterized as refractory. Furthermore, 28 of the 66 AML mutations analyzed had distinct metabolites and/or lipids associated with it, distinct pathway enrichment (Figure 1), and distinct patterns in lipid classes, length, and saturation levels (supplemental Figure 2). Intriguingly, some of these findings align with known AML biology such as the increase in 2-hydroxyglutarate in IDH1 mutant AML samples26 (Figure 1A) and glutamate metabolism in FLT3-ITD (internal tandem duplication) mutant samples27 (Figure 1C). These findings demonstrate that our circulating metabolome resource has the potential to enable the discovery of novel metabolic frameworks across AML subtypes. There has been growing interest in uncovering lipidomic changes in AML.28 Although there are excellent resources for identifying biologically relevant lipids29,30 using high yield input techniques,22 lipid metabolism research still lags behind metabolomics and other omics in availability of comprehensive tools and databases. Thus, the data published here can help fill this knowledge gap as an AML plasma lipidome resource.
Next, to build a model predictive of patient survival, machine learning models were trained to identify refractory AML using circulating lipid levels, with and without diagnostically relevant clinical features used to generate other prognostic signatures (age, cytogenetic risk, European Leukemia Network risk, white blood cell count, and AML diagnosis31), as well as metabolite data and clinical features alone (supplemental Table 5). The lipid signature developed using GBC had exceptional accuracy categorizing therapeutic response. Lipids alone (area under the curve [AUC], 0.94) and lipids with clinical features (AUC, 0.93) performed nearly identically, surpassing the performance of metabolites (AUC, 0.56) and clinical features alone (AUC, 0.59; Figure 2A), suggesting that lipids alone could be prospective biomarkers for patient outcomes.
Circulating lipid signature can predict patient survival. To predict patient outcomes, a machine learning model was developed for lipidomics data, with and without the addition of clinical parameters. After training the algorithm on 80% of the PMH data set, a holdout testing subset (20%) of the PMH data set was used to evaluate the model performance. (A) Lipids with clinical parameters compared similarly to lipids alone, and lipids vastly outperform models developed for metabolites or clinical parameters alone. (B) The testing PMH data subset was also used to evaluate patient survival based on the lipid signature derived from the machine learning model. Overall patient survival, based on the abundance of the top lipid in the lipid signature alone; SM (d44:1) outcomes in the entire PMH data set (C) and the entire UC data set (D).
Circulating lipid signature can predict patient survival. To predict patient outcomes, a machine learning model was developed for lipidomics data, with and without the addition of clinical parameters. After training the algorithm on 80% of the PMH data set, a holdout testing subset (20%) of the PMH data set was used to evaluate the model performance. (A) Lipids with clinical parameters compared similarly to lipids alone, and lipids vastly outperform models developed for metabolites or clinical parameters alone. (B) The testing PMH data subset was also used to evaluate patient survival based on the lipid signature derived from the machine learning model. Overall patient survival, based on the abundance of the top lipid in the lipid signature alone; SM (d44:1) outcomes in the entire PMH data set (C) and the entire UC data set (D).
The GBC-predicted high-risk lipid signature was also able to stratify patient overall survival (Figure 2B). Supporting this finding, multivariable modeling demonstrated based on concordance scoring (c-score) that the addition of the lipid signature to the best clinical metrics of outcomes improved outcome predictions vs (c-score, 0.789) clinical parameters alone (c-score, 0.711; supplemental Table 6), demonstrating the prognostic value of circulating lipids in this patient cohort.
Finally, to identify individual lipid survival biomarkers, we evaluated the top lipids in the GBC lipid signature by univariate Cox regression and Kaplan-Meier curve analysis. Although many lipids were significantly associated with survival, SM (d44:1) had the greatest capability to predict overall survival in the PMH data set (Figure 2C). To validate the utility of lipids to predict survival across AML, a distinct validation data set consisting of 67 diagnosis bone marrow plasma samples was obtained from an unrelated clinical center where patients received 7+3 as well as other induction therapies (supplemental Table 7). Although this independent data set (the University of Colorado [UC] data set) was obtained from bone marrow and not peripheral blood, overall patient survival in the UC data set was significantly stratified based on SM (d44:1; Figure 2D). However, the profile of the lipidome within the UC data set was quite different from the PMH data set, suggesting that the site of circulating lipidome collection may be influenced by the local microenvironment.
Sphingolipids have previously been established as important in AML biology and can be targeted to eradicate the disease.32-35 Indeed, cellular levels of SMs are also prognostic of survival in AML, with high levels of cellular SM being associated with worse outcomes.36 Our study shows that low SM (d44:1) in plasma is independently associated with worse outcomes (supplemental Figure 3), suggesting a critical role of SM transport and metabolism relating to disease aggressiveness. Thus, SM (d44:1) and related lipid species should be investigated as potential circulating biomarkers of AML survival with the potential to improve decision-making for health care providers.
Biomarker research in AML has primarily focused on mutation screening and gene expression, of which many have highlighted lipid signatures as highly useful for stratifying patients with AML.37-40 Lipids from peripheral blood represent a potentially underused, noninvasive, and abundant biomarker. In some solid tumor cancers, lipid biomarkers have already been used for diagnosis, prognosis, and risk stratification.14,15,41,42 In AML, although lipids can stratify healthy individuals from patients,16,19 their association with prognosis and outcomes have only been suggested.20,21,36,43,44 Expanding on this area of research, here, we propose that circulating lipids are more deeply connected with AML’s underlying biology and that lipids in circulation are associated with distinct molecular features of AML. Although further validation is required, we revealed that some AML drivers such as TP53, RAD21, or PTPN11 have detectable and unique presentation of lipids but not metabolites in circulation. Thus, these AML subtypes might have previously unidentified effects on local and global metabolism and may be more sensitive to lipid metabolism interventions. Additional research will be required to identify the underlying mechanism driving these metabolite and lipid patterns, which could include metabolite release from intact leukemic cells, lysed leukemic cells, or extracellular vesicles. Furthermore, using advanced lipidomics techniques, we have detected nearly 2000 lipid species, far surpassing what has previously been published on circulating lipids in AML, making this work a highly useful resource for studying unique molecular phenotypes as well as AML lipid metabolism overall.
Importantly, the critical role of lipid metabolism in AML biology is demonstrated by the ability of circulating lipids to predict overall survival. The introduction of circulating lipids as potential biomarkers may lead to novel treatment strategies for patients and improve outcomes. However, additional clinical validation will be necessary to fully translate our discoveries.
Acknowledgments
The authors thank the Leukemia Tissue Bank (Princess Margaret Cancer Centre) for providing the primary AML samples; Jill Flewelling, Sally DeSilva, and Jess Widner for coordination and administrative support; Fabia Gamboni for assistance with omics data acquisition; the laboratory of K.H., particularly Saeer Adeel, for project guidance; and Craig Jordan for his thoughtful feedback.
This study was funded, in part, by a Medical Biophysics Excellence Ontario Student Opportunity Trust Fund Award (C.O.); a Sona Noran Pancha Graduate Award (C.O.); a David Rae Graduate Student Scholarship (C.O.); an Ontario Graduate Scholarship (C.O.); Blood Cancer United (formerly Leukemia and Lymphoma Society) Blood Cancer Discovery grant 8035-23 (C.L.J.); the Elsa Pardee Foundation (C.L.J.); Rally Foundation grant 24CDN01 (C.L.J.); V Foundation grant V2024-012 (C.L.J.); Alex’s Lemonade Stand Foundation grant 1330583 (C.L.J.); a Cincinnati Children’s Hospital Medical Center Trustee Award (C.L.J.); the Canadian Institute of Health Research (C.L.J. and K.H.); the Princess Margaret Cancer Center, the Princess Margaret Cancer Foundation, and the Ontario Ministry of Health (K.H., A.T., M.D.M., A.A., B.S., and C.L.J.); University of Colorado Cancer Center Support grant (P30CA046934 [A.D.]); National Cancer Institute, National Institutes of Health grant 1F31CA250361-01 (R.C.-H.); and the Blood Cancer United (formerly the Leukemia and Lymphoma Society) Career Development Program Scholar in Clinical Research Achievement Award (D.A.P.).
Authorship
Contribution: C.O. designed and performed the research, analyzed and interpreted data, and wrote the manuscript; N.N. directed machine learning analysis, analyzed and interpreted data, and wrote the manuscript; A.T. performed metabolomic and lipidomic experiments, analyzed data, and wrote the manuscript; R.C.-H. performed metabolomics experiments, collected, analyzed, and interpreted metabolomics data, and wrote the manuscript; A.A. and M.D.M. provided acute myeloid leukemia (AML) specimens, advised on the design of the research study, and wrote the manuscript; T.M. and A.K. collected clinical patient data and wrote the manuscript; B.S. and D.A.P. provided AML specimens and wrote the manuscript; K.H. designed and directed research, analyzed and interpreted data, and wrote the manuscript; and S.K., J.A.R., A.D., and C.L.J. designed research, analyzed and interpreted metabolomics and lipidomics data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Courtney L. Jones, Cincinnati Children's Hospital Medical Center, 3333 Burnet Ave MLC 7013, Cincinnati, OH 45229; email: courtney.jones@cchmc.org; Angelo D'Alessandro, University of Colorado Anschutz Medical Campus, 12801 E 17th Ave, Aurora, CO 80045 80045; email: angelo.dalessandro@cuanschutz.edu; Julie A. Reisz, University of Colorado Anschutz Medical Campus, 12801 E 17th Ave, Aurora, CO 80045; email: julie.haines@cuanschutz.edu; and Sushant Kumar, Princess Margaret Cancer Research Centre, 101 College St, Room 12-706, Toronto, ON M5G 1L7 1L7, Canada; email: sushant.kumar@uhn.ca.
References
Author notes
C.O. and N.N. contributed equally to this study.
All raw data metabolite and lipid data are available in the supplemental Files and in the shinyapp (available at https://plasmalipids.shinyapps.io/AML_circulating_metabolites/).
Additional data or information is available on request from corresponding author Courtney L. Jones (courtney.jones@cchmc.org).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.