TO THE EDITOR:
Sickle cell trait (HbAS) is a common genetic carrier state found in ∼3 million individuals in the United States, who are predominantly from the Black race.1 Although HbAS was long considered benign, recent evidence has demonstrated strong clinical associations between HbAS and an increased risk for chronic kidney disease (CKD) and venous thromboembolisms (VTE).2,3 Consequently, the American Society of Hematology has called for further research to better understand these clinical associations in HbAS, including genetic research.4 MicroRNAs are a crucial component of genetic research due to their role in modulating gene expression via messenger RNA regulation.5,6 By modulating gene expression, microRNAs regulate the activity of various signaling pathways involved in disease pathogenesis.5 Consequently, the study of circulating microRNAs may provide insights into various pathophysiologies.
To better understand whether the presence of HbAS may be an important factor in the modulation of gene expression, we sought to compare the differential expression of circulating microRNAs in individuals with HbAS with the normal hemoglobin phenotype (HbAA). We hypothesized that individuals with HbAS would have a significant differential expression of several microRNAs compared with HbAA.
This was a cross-sectional study using data and plasma samples from the Mass General Brigham (MGB; formerly called Partners Healthcare) Biobank collected between 2005 and 2018. The MGB Biobank comprises samples voluntarily obtained with informed consent from individuals at MGB-affiliated hospitals and has been described in previous studies.7 MGB Biobank specimens are linked to clinical data from the electronic medical records of affiliated hospitals. The MGB and University of Texas Southwestern institutional review boards approved this study (University of Texas Southwestern institutional review board number STU2020-1053).
We applied the following inclusion criteria at baseline (the time of sample collection): self-identified Black race, age ≥18 years, an estimated glomerular filtration rate (eGFR) >60 mL/min, and the presence of urine protein studies. The following exclusion criteria were applied at baseline: age ≥65 years, the presence of any comorbidity (eg, diabetes mellitus, hypertension, cardiovascular disease, CKD, cancer, transplantation, and autoimmune disease), or proteinuria. Matching was performed (1:1) on age ±5 years, sex, and eGFR ±20 mL/min. HbAS and HbAA were confirmed by a hemoglobin electrophoresis test interpreted by a pathologist. Frozen plasma samples (0.5 mL per patient) were sent to LC Sciences, Houston,8-10 for processing. Total RNA was extracted using TRIzol reagent, following the manufacturer's procedure. The quality and quantity of total RNA were analyzed by Bioanalyzer 2100, with a RNA Integrity number >7.0. Approximately 1 μg of total RNA was used to prepare a small RNA library using TruSeq Small RNA Sample Prep Kits, following the manufacturer's protocol. Subsequently, single-end sequencing (50 base pairs) was performed on an Illumina HiSeq 2500 at LC Sciences, following the vendor's recommended protocol. Raw reads were subjected to an in-house program, ACGT101-miR, to remove adapter dimers, junk, low complexity, common RNA families (ribosomal RNA, transfer RNA, small nuclear RNA [snRNA], and small nucleolar RNA [snoRNA]), and repeats. Subsequently, unique sequences with lengths ranging from 18 to 26 nucleotides were mapped to specific species precursors in miRBase 22.0 by Basic Local Alignment Search Tool (BLAST) search to identify known microRNAs and novel 3p- and 5p-derived microRNAs.
Normalization of microRNA sequence counts in each sample was achieved by dividing the counts by a library size parameter of the corresponding sample. The library size parameter is a median value of the ratio between the counts of a specific sample and a pseudoreference sample. A count number in the pseudoreference sample is the count geometric mean across all samples.
To predict the genes targeted by the most abundant microRNAs, 2 computational target prediction algorithms (TargetScan and miRanda) were used to identify microRNA binding sites. Finally, the data predicted by both algorithms were combined, and the overlaps were calculated. The Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic signaling pathways of these most abundant microRNAs were also annotated. Significant KEGG metabolic pathways were calculated by a hypergeometric equation. Those KEGG pathways with a P value <.05 were defined as significant KEGG metabolic pathways.
Differential expression of microRNAs based on normalized deep-sequencing counts were analyzed using the nonparametric Wilcoxon rank-sum test. The significance threshold was set to <.05 for each test.
We identified 200 individuals with hemoglobin electrophoresis results and available plasma samples (182 with HbAA and 18 with HbAS). Eighty with HbAA and 12 individuals with HbAS remained after simultaneously applying inclusion and exclusion criteria. After matching, 12 with HbAA were included. Compared with HbAA, individuals with HbAS had a lower baseline eGFR (101 ± 19 vs 116 ± 17 mL/min; Figure 1).
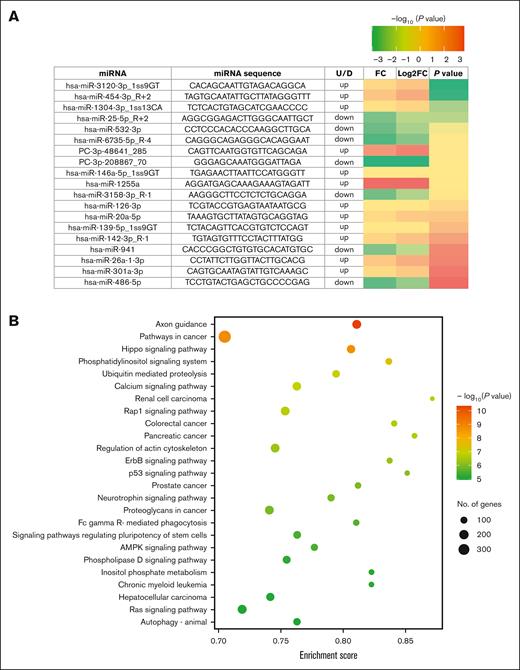
Differential expression of miRNA in HbAS. (A) Heat map of differentially expressed plasma miRNAs in sickle cell anemia compared with the normal HbAA. (B) Bubble plot of significant KEGG pathway enrichment analysis (top 25). Characteristics of the HbAS cohort (n = 12) were as follows: mean age, 41 ± 7 years; 42% female; and mean eGFR, 101 ± 19 mL/min. The normal HbAA cohort (n = 12) had a mean age of 42 ± 8 years, 42% female, and a mean eGFR of 116 ± 17 mL/min. All 24 participants self-identified as Black race, were not on any chronic medications, and had no history of comorbidities. FC, fold change; miRNA, microRNA; U/D, up/down.
Differential expression of miRNA in HbAS. (A) Heat map of differentially expressed plasma miRNAs in sickle cell anemia compared with the normal HbAA. (B) Bubble plot of significant KEGG pathway enrichment analysis (top 25). Characteristics of the HbAS cohort (n = 12) were as follows: mean age, 41 ± 7 years; 42% female; and mean eGFR, 101 ± 19 mL/min. The normal HbAA cohort (n = 12) had a mean age of 42 ± 8 years, 42% female, and a mean eGFR of 116 ± 17 mL/min. All 24 participants self-identified as Black race, were not on any chronic medications, and had no history of comorbidities. FC, fold change; miRNA, microRNA; U/D, up/down.
Nineteen differentially expressed plasma microRNAs were identified in HbAS (12 increased and 7 reduced abundance) compared with HbAA (Figure 1).
The differentially expressed microRNAs in HbAS were linked to 100 target genes (Table 1) and mapped to 99 significant metabolic signaling pathways, based on gene enrichment (Figure 1).
Gene target prediction of differentially expressed microRNAs
| Gene annotation (symbol) . | microRNA ID . | TargetScan score . | miRanda energy score . |
|---|---|---|---|
| ADP ribosylation factor 5 (ARF5) | hsa-miR-3120-3p_1ss9GT | 87 | –13.5 |
| FKBP prolyl isomerase 4 (FKBP4) | PC-3p-208867_70 | 93 | –19.72 |
| FKBP prolyl isomerase 4 (FKBP4) | hsa-miR-1255a | 73 | –44.01 |
| cytochrome p450 family 26 subfamily B member 1 (CYP26B1) | hsa-miR-1255a | 50 | –18.09 |
| cytochrome p450 family 26 subfamily B member 1 (CYP26B1) | hsa-miR-1304-3p_1ss13CA | 82 | –20.50 |
| cytochrome p450 family 26 subfamily B member 1 (CYP26B1) | hsa-miR-20a-5p | 66 | –30.95 |
| cytochrome p450 family 26 subfamily B member 1 (CYP26B1) | hsa-miR-3120-3p_1ss9GT | 81 | –12.45 |
| cytochrome p450 family 26 subfamily B member 1 (CYP26B1) | hsa-miR-532-3p | 63 | –58.52 |
| NADH-ubiquinone oxidoreductase complex assembly factor 7 (NDUFAF7) | hsa-miR-941 | 99 | –20.54 |
| alpha-l-fucosidase 2 (FUCA2) | PC-3p-208867_70 | 72 | –12.18 |
| alpha-l-fucosidase 2 (FUCA2) | hsa-miR-1304-3p_1ss13CA | 81 | –18.62 |
| alpha-l-fucosidase 2 (FUCA2) | hsa-miR-139-5p_1ss9GT | 79 | –27.04 |
| alpha-l-fucosidase 2 (FUCA2) | hsa-miR-142-3p_R-1 | 98 | –19.16 |
| alpha-l-fucosidase 2 (FUCA2) | hsa-miR-20a-5p | 74 | –30.40 |
| heparan sulfate glucosamine 3-sulfotransferase 1 (HS3ST1) | PC-3p-208867_70 | 99 | –11.24 |
| heparan sulfate glucosamine3-sulfotransferase1 (HS3ST1) | hsa-miR-1304-3p_1ss13CA | 77 | –15.99 |
| heparan sulfate glucosamine3-sulfotransferase1 (HS3ST1) | hsa-miR-26a-1-3p | 89 | –25.90 |
| heparan sulfate glucosamine3-sulfotransferase1 (HS3ST1) | hsa-miR-3120-3p_1ss9GT | 72 | –16.25 |
| semaphorin 3F (SEMA3F) | hsa-miR-25-5p_R+2 | 94 | –18.49 |
| semaphorin 3F (SEMA3F) | hsa-miR-3158-3p_R-1 | 74 | –23.37 |
| CF transmembrane conductance regulator (CFTR) | PC-3p-48641_285 | 86 | –14.45 |
| cytochrome p450 family 51 subfamily A member 1 (CYP51A1) | hsa-miR-1304-3p_1ss13CA | 96 | –12.87 |
| ubiquitin specific peptidase 28 (USP28) | hsa-miR-1304-3p_1ss13CA | 93 | –15.17 |
| ubiquitin specific peptidase 28 (USP28) | hsa-miR-301a-3p | 95 | –15.63 |
| ubiquitin specific peptidase 28 (USP28) | hsa-miR-454-3p_R+2 | 93 | –13.24 |
| sperm tailPG-richrepeat containing 1 (STPG1) | PC-3p-208867_70 | 58 | –19.70 |
| sperm tailPG-richrepeat containing 1 (STPG1) | hsa-miR-532-3p | 94 | –21.41 |
| solute carrier family 7 member 2 (SLC7A2) | PC-3p-48641_285 | 61 | –16.32 |
| solute carrier family 7 member 2 (SLC7A2) | hsa-miR-1304-3p_1ss13CA | 94 | –56.97 |
| solute carrier family 7 member 2 (SLC7A2) | hsa-miR-146a-5p_1ss9GT | 61 | –12.98 |
| solute carrier family 7 member 2 (SLC7A2) | hsa-miR-20a-5p | 60 | –15.71 |
| solute carrier family 7 member 2 (SLC7A2) | hsa-miR-26a-1-3p | 69 | –24.58 |
| solute carrier family 7 member 2 (SLC7A2) | hsa-miR-3120-3p_1ss9GT | 57 | –114.84 |
| solute carrier family 7 member 2 (SLC7A2) | hsa-miR-486-5p | 87 | –17.37 |
| solute carrier family 7 member 2 (SLC7A2) | hsa-miR-532-3p | 90 | –40.46 |
| heat shock protein family B (small) member 6 (HSPB6) | hsa-miR-25-5p_R+2 | 89 | –38.41 |
| heat shock protein family B (small) member 6 (HSPB6) | hsa-miR-6735-5p_R-4 | 90 | –29.25 |
| pyruvate dehydrogenase kinase 4 (PDK4) | hsa-miR-532-3p | 83 | –18.45 |
| USH1 protein network component harmonin (USH1C) | hsa-miR-142-3p_R-1 | 78 | –19.36 |
| USH1 protein network component harmonin (USH1C) | hsa-miR-6735-5p_R-4 | 75 | –31.63 |
| RAS likeproto-oncogeneA (RALA) | hsa-miR-139-5p_1ss9GT | 60 | –11.26 |
| RAS likeproto-oncogeneA (RALA) | hsa-miR-301a-3p | 89 | –14.09 |
| RAS likeproto-oncogeneA (RALA) | hsa-miR-454-3p_R+2 | 89 | –17.18 |
| RAS likeproto-oncogeneA (RALA) | hsa-miR-532-3p | 77 | –28.50 |
| B cell receptor associated protein 29 (BCAP29) | hsa-miR-301a-3p | 59 | –26.11 |
| B cell receptor associated protein 29 (BCAP29) | hsa-miR-454-3p_R+2 | 63 | –26.93 |
| B cell receptor associated protein 29 (BCAP29) | hsa-miR-6735-5p_R-4 | 64 | –73.17 |
| BAR/IMD domain containing adaptor protein 2 like 1 (BAIAP2L1) | hsa-miR-6735-5p_R-4 | 57 | –65.25 |
| RNA polymerase II associated protein 3 (RPAP3) | PC-3p-48641_285 | 95 | –12.57 |
| RNA polymerase II associated protein 3 (RPAP3) | hsa-miR-1255a | 51 | –18.99 |
| RNA polymerase II associated protein 3 (RPAP3) | hsa-miR-139-5p_1ss9GT | 71 | –19.47 |
| uridine phosphorylase 2 (UPP2) | hsa-miR-1255a | 99 | –11.09 |
| uridine phosphorylase 2 (UPP2) | hsa-miR-146a-5p_1ss9GT | 99 | –12.59 |
| uridine phosphorylase 2 (UPP2) | hsa-miR-26a-1-3p | 50 | –20.71 |
| homeobox A11 (HOXA11) | hsa-miR-532-3p | 79 | –17.59 |
| C-X3-Cmotif chemokine ligand 1 (CX3CL1) | hsa-miR-20a-5p | 55 | –13.69 |
| C-X3-Cmotif chemokine ligand 1 (CX3CL1) | hsa-miR-6735-5p_R-4 | 71 | –150.88 |
| CEA cell adhesion molecule 7 (CEACAM7) | hsa-miR-3120-3p_1ss9GT | 55 | –19.40 |
| CEA cell adhesion molecule 7 (CEACAM7) | hsa-miR-6735-5p_R-4 | 94 | –17.76 |
| rhomboid domain containing 2 (RHBDD2) | hsa-miR-6735-5p_R-4 | 86 | –48.16 |
| oxysterol binding protein like 7 (OSBPL7) | hsa-miR-1304-3p_1ss13CA | 55 | –23.05 |
| oxysterol binding protein like 7 (OSBPL7) | hsa-miR-6735-5p_R-4 | 62 | –52.65 |
| NADH: ubiquinone oxidoreductase subunit AB1 (NDUFAB1) | hsa-miR-1304-3p_1ss13CA | 77 | –20.64 |
| integrin subunit alpha 3 (ITGA3) | hsa-miR-3120-3p_1ss9GT | 74 | –14.05 |
| cryptochrome circadian regulator 1 (CRY1) | hsa-miR-146a-5p_1ss9GT | 91 | –20.38 |
| cryptochrome circadian regulator 1 (CRY1) | hsa-miR-301a-3p | 86 | –14.94 |
| cryptochrome circadian regulator 1 (CRY1) | hsa-miR-454-3p_R+2 | 82 | –10.37 |
| STARD3 N-terminal like (STARD3NL) | PC-3p-48641_285 | 85 | –22.34 |
| STARD3 N-terminal like (STARD3NL) | hsa-miR-1304-3p_1ss13CA | 97 | –15.16 |
| CD74 molecule (CD74) | PC-3p-208867_70 | 60 | –19.51 |
| CD74 molecule (CD74) | hsa-miR-1255a | 85 | –43.07 |
| CD74 molecule (CD74) | hsa-miR-532-3p | 83 | –17.95 |
| synaptophysin like 1 (SYPL1) | hsa-miR-142-3p_R-1 | 83 | –14.84 |
| synaptophysin like 1 (SYPL1) | hsa-miR-3120-3p_1ss9GT | 62 | –10.73 |
| synaptophysin like 1 (SYPL1) | hsa-miR-486-5p | 87 | –12.65 |
| RAN binding protein 9 (RANBP9) | hsa-miR-532-3p | 71 | –23.39 |
| pleckstrin homology and RhoGEF domain containing G6 (PLEKHG6) | hsa-miR-532-3p | 58 | –24.82 |
| pleckstrin homology and RhoGEF domain containing G6 (PLEKHG6) | hsa-miR-6735-5p_R-4 | 76 | –44.22 |
| vacuolar protein sorting 13 homolog D (VPS13D) | PC-3p-48641_285 | 67 | –18.56 |
| vacuolar protein sorting 13 homolog D (VPS13D) | hsa-miR-301a-3p | 98 | –17.82 |
| vacuolar protein sorting 13 homolog D (VPS13D) | hsa-miR-454-3p_R+2 | 97 | –14.07 |
| tetraspanin 9 (TSPAN9) | hsa-miR-1304-3p_1ss13CA | 51 | –36.07 |
| tetraspanin 9 (TSPAN9) | hsa-miR-6735-5p_R-4 | 50 | –124.32 |
| glutaminyl-peptidecyclotransferase-like(QPCTL) | PC-3p-208867_70 | 72 | –13.72 |
| glutaminyl-peptidecyclotransferase-like(QPCTL) | hsa-miR-1304-3p_1ss13CA | 59 | –24.63 |
| glutaminyl-peptidecyclotransferase-like(QPCTL) | hsa-miR-25-5p_R+2 | 83 | –21.50 |
| HIVEP zinc finger 2 (HIVEP2) | hsa-miR-301a-3p | 94 | –11.43 |
| HIVEP zinc finger 2 (HIVEP2) | hsa-miR-454-3p_R+2 | 94 | –15.18 |
| HIVEP zinc finger 2 (HIVEP2) | hsa-miR-532-3p | 76 | –47.97 |
| ubiquitin protein ligase E3 component n-recognin 7 (UBR7) | hsa-miR-139-5p_1ss9GT | 57 | –11.59 |
| ubiquitin protein ligase E3 component n-recognin 7 (UBR7) | hsa-miR-301a-3p | 88 | –27.41 |
| ubiquitin protein ligase E3 component n-recognin 7 (UBR7) | hsa-miR-454-3p_R+2 | 89 | –27.50 |
| indolethylamine N-methyltransferase (INMT) | hsa-miR-486-5p | 55 | –15.57 |
| abhydrolase domain containing 5, lysophosphatidic acid acyltransferase (ABHD5) | hsa-miR-146a-5p_1ss9GT | 94 | –18.01 |
| Gprotein-coupledreceptor class C group 5 member A (GPRC5A) | hsa-miR-1255a | 94 | –51.69 |
| Gprotein-coupledreceptor class C group 5 member A (GPRC5A) | hsa-miR-142-3p_R-1 | 63 | –34.07 |
| Gprotein-coupledreceptor class C group 5 member A (GPRC5A) | hsa-miR-3120-3p_1ss9GT | 55 | –31.85 |
| Gprotein-coupledreceptor class C group 5 member A (GPRC5A) | hsa-miR-532-3p | 93 | –50.65 |
| cytochrome c oxidase assembly homolog COX15 (COX15) | PC-3p-208867_70 | 83 | –17.63 |
| Gene annotation (symbol) . | microRNA ID . | TargetScan score . | miRanda energy score . |
|---|---|---|---|
| ADP ribosylation factor 5 (ARF5) | hsa-miR-3120-3p_1ss9GT | 87 | –13.5 |
| FKBP prolyl isomerase 4 (FKBP4) | PC-3p-208867_70 | 93 | –19.72 |
| FKBP prolyl isomerase 4 (FKBP4) | hsa-miR-1255a | 73 | –44.01 |
| cytochrome p450 family 26 subfamily B member 1 (CYP26B1) | hsa-miR-1255a | 50 | –18.09 |
| cytochrome p450 family 26 subfamily B member 1 (CYP26B1) | hsa-miR-1304-3p_1ss13CA | 82 | –20.50 |
| cytochrome p450 family 26 subfamily B member 1 (CYP26B1) | hsa-miR-20a-5p | 66 | –30.95 |
| cytochrome p450 family 26 subfamily B member 1 (CYP26B1) | hsa-miR-3120-3p_1ss9GT | 81 | –12.45 |
| cytochrome p450 family 26 subfamily B member 1 (CYP26B1) | hsa-miR-532-3p | 63 | –58.52 |
| NADH-ubiquinone oxidoreductase complex assembly factor 7 (NDUFAF7) | hsa-miR-941 | 99 | –20.54 |
| alpha-l-fucosidase 2 (FUCA2) | PC-3p-208867_70 | 72 | –12.18 |
| alpha-l-fucosidase 2 (FUCA2) | hsa-miR-1304-3p_1ss13CA | 81 | –18.62 |
| alpha-l-fucosidase 2 (FUCA2) | hsa-miR-139-5p_1ss9GT | 79 | –27.04 |
| alpha-l-fucosidase 2 (FUCA2) | hsa-miR-142-3p_R-1 | 98 | –19.16 |
| alpha-l-fucosidase 2 (FUCA2) | hsa-miR-20a-5p | 74 | –30.40 |
| heparan sulfate glucosamine 3-sulfotransferase 1 (HS3ST1) | PC-3p-208867_70 | 99 | –11.24 |
| heparan sulfate glucosamine3-sulfotransferase1 (HS3ST1) | hsa-miR-1304-3p_1ss13CA | 77 | –15.99 |
| heparan sulfate glucosamine3-sulfotransferase1 (HS3ST1) | hsa-miR-26a-1-3p | 89 | –25.90 |
| heparan sulfate glucosamine3-sulfotransferase1 (HS3ST1) | hsa-miR-3120-3p_1ss9GT | 72 | –16.25 |
| semaphorin 3F (SEMA3F) | hsa-miR-25-5p_R+2 | 94 | –18.49 |
| semaphorin 3F (SEMA3F) | hsa-miR-3158-3p_R-1 | 74 | –23.37 |
| CF transmembrane conductance regulator (CFTR) | PC-3p-48641_285 | 86 | –14.45 |
| cytochrome p450 family 51 subfamily A member 1 (CYP51A1) | hsa-miR-1304-3p_1ss13CA | 96 | –12.87 |
| ubiquitin specific peptidase 28 (USP28) | hsa-miR-1304-3p_1ss13CA | 93 | –15.17 |
| ubiquitin specific peptidase 28 (USP28) | hsa-miR-301a-3p | 95 | –15.63 |
| ubiquitin specific peptidase 28 (USP28) | hsa-miR-454-3p_R+2 | 93 | –13.24 |
| sperm tailPG-richrepeat containing 1 (STPG1) | PC-3p-208867_70 | 58 | –19.70 |
| sperm tailPG-richrepeat containing 1 (STPG1) | hsa-miR-532-3p | 94 | –21.41 |
| solute carrier family 7 member 2 (SLC7A2) | PC-3p-48641_285 | 61 | –16.32 |
| solute carrier family 7 member 2 (SLC7A2) | hsa-miR-1304-3p_1ss13CA | 94 | –56.97 |
| solute carrier family 7 member 2 (SLC7A2) | hsa-miR-146a-5p_1ss9GT | 61 | –12.98 |
| solute carrier family 7 member 2 (SLC7A2) | hsa-miR-20a-5p | 60 | –15.71 |
| solute carrier family 7 member 2 (SLC7A2) | hsa-miR-26a-1-3p | 69 | –24.58 |
| solute carrier family 7 member 2 (SLC7A2) | hsa-miR-3120-3p_1ss9GT | 57 | –114.84 |
| solute carrier family 7 member 2 (SLC7A2) | hsa-miR-486-5p | 87 | –17.37 |
| solute carrier family 7 member 2 (SLC7A2) | hsa-miR-532-3p | 90 | –40.46 |
| heat shock protein family B (small) member 6 (HSPB6) | hsa-miR-25-5p_R+2 | 89 | –38.41 |
| heat shock protein family B (small) member 6 (HSPB6) | hsa-miR-6735-5p_R-4 | 90 | –29.25 |
| pyruvate dehydrogenase kinase 4 (PDK4) | hsa-miR-532-3p | 83 | –18.45 |
| USH1 protein network component harmonin (USH1C) | hsa-miR-142-3p_R-1 | 78 | –19.36 |
| USH1 protein network component harmonin (USH1C) | hsa-miR-6735-5p_R-4 | 75 | –31.63 |
| RAS likeproto-oncogeneA (RALA) | hsa-miR-139-5p_1ss9GT | 60 | –11.26 |
| RAS likeproto-oncogeneA (RALA) | hsa-miR-301a-3p | 89 | –14.09 |
| RAS likeproto-oncogeneA (RALA) | hsa-miR-454-3p_R+2 | 89 | –17.18 |
| RAS likeproto-oncogeneA (RALA) | hsa-miR-532-3p | 77 | –28.50 |
| B cell receptor associated protein 29 (BCAP29) | hsa-miR-301a-3p | 59 | –26.11 |
| B cell receptor associated protein 29 (BCAP29) | hsa-miR-454-3p_R+2 | 63 | –26.93 |
| B cell receptor associated protein 29 (BCAP29) | hsa-miR-6735-5p_R-4 | 64 | –73.17 |
| BAR/IMD domain containing adaptor protein 2 like 1 (BAIAP2L1) | hsa-miR-6735-5p_R-4 | 57 | –65.25 |
| RNA polymerase II associated protein 3 (RPAP3) | PC-3p-48641_285 | 95 | –12.57 |
| RNA polymerase II associated protein 3 (RPAP3) | hsa-miR-1255a | 51 | –18.99 |
| RNA polymerase II associated protein 3 (RPAP3) | hsa-miR-139-5p_1ss9GT | 71 | –19.47 |
| uridine phosphorylase 2 (UPP2) | hsa-miR-1255a | 99 | –11.09 |
| uridine phosphorylase 2 (UPP2) | hsa-miR-146a-5p_1ss9GT | 99 | –12.59 |
| uridine phosphorylase 2 (UPP2) | hsa-miR-26a-1-3p | 50 | –20.71 |
| homeobox A11 (HOXA11) | hsa-miR-532-3p | 79 | –17.59 |
| C-X3-Cmotif chemokine ligand 1 (CX3CL1) | hsa-miR-20a-5p | 55 | –13.69 |
| C-X3-Cmotif chemokine ligand 1 (CX3CL1) | hsa-miR-6735-5p_R-4 | 71 | –150.88 |
| CEA cell adhesion molecule 7 (CEACAM7) | hsa-miR-3120-3p_1ss9GT | 55 | –19.40 |
| CEA cell adhesion molecule 7 (CEACAM7) | hsa-miR-6735-5p_R-4 | 94 | –17.76 |
| rhomboid domain containing 2 (RHBDD2) | hsa-miR-6735-5p_R-4 | 86 | –48.16 |
| oxysterol binding protein like 7 (OSBPL7) | hsa-miR-1304-3p_1ss13CA | 55 | –23.05 |
| oxysterol binding protein like 7 (OSBPL7) | hsa-miR-6735-5p_R-4 | 62 | –52.65 |
| NADH: ubiquinone oxidoreductase subunit AB1 (NDUFAB1) | hsa-miR-1304-3p_1ss13CA | 77 | –20.64 |
| integrin subunit alpha 3 (ITGA3) | hsa-miR-3120-3p_1ss9GT | 74 | –14.05 |
| cryptochrome circadian regulator 1 (CRY1) | hsa-miR-146a-5p_1ss9GT | 91 | –20.38 |
| cryptochrome circadian regulator 1 (CRY1) | hsa-miR-301a-3p | 86 | –14.94 |
| cryptochrome circadian regulator 1 (CRY1) | hsa-miR-454-3p_R+2 | 82 | –10.37 |
| STARD3 N-terminal like (STARD3NL) | PC-3p-48641_285 | 85 | –22.34 |
| STARD3 N-terminal like (STARD3NL) | hsa-miR-1304-3p_1ss13CA | 97 | –15.16 |
| CD74 molecule (CD74) | PC-3p-208867_70 | 60 | –19.51 |
| CD74 molecule (CD74) | hsa-miR-1255a | 85 | –43.07 |
| CD74 molecule (CD74) | hsa-miR-532-3p | 83 | –17.95 |
| synaptophysin like 1 (SYPL1) | hsa-miR-142-3p_R-1 | 83 | –14.84 |
| synaptophysin like 1 (SYPL1) | hsa-miR-3120-3p_1ss9GT | 62 | –10.73 |
| synaptophysin like 1 (SYPL1) | hsa-miR-486-5p | 87 | –12.65 |
| RAN binding protein 9 (RANBP9) | hsa-miR-532-3p | 71 | –23.39 |
| pleckstrin homology and RhoGEF domain containing G6 (PLEKHG6) | hsa-miR-532-3p | 58 | –24.82 |
| pleckstrin homology and RhoGEF domain containing G6 (PLEKHG6) | hsa-miR-6735-5p_R-4 | 76 | –44.22 |
| vacuolar protein sorting 13 homolog D (VPS13D) | PC-3p-48641_285 | 67 | –18.56 |
| vacuolar protein sorting 13 homolog D (VPS13D) | hsa-miR-301a-3p | 98 | –17.82 |
| vacuolar protein sorting 13 homolog D (VPS13D) | hsa-miR-454-3p_R+2 | 97 | –14.07 |
| tetraspanin 9 (TSPAN9) | hsa-miR-1304-3p_1ss13CA | 51 | –36.07 |
| tetraspanin 9 (TSPAN9) | hsa-miR-6735-5p_R-4 | 50 | –124.32 |
| glutaminyl-peptidecyclotransferase-like(QPCTL) | PC-3p-208867_70 | 72 | –13.72 |
| glutaminyl-peptidecyclotransferase-like(QPCTL) | hsa-miR-1304-3p_1ss13CA | 59 | –24.63 |
| glutaminyl-peptidecyclotransferase-like(QPCTL) | hsa-miR-25-5p_R+2 | 83 | –21.50 |
| HIVEP zinc finger 2 (HIVEP2) | hsa-miR-301a-3p | 94 | –11.43 |
| HIVEP zinc finger 2 (HIVEP2) | hsa-miR-454-3p_R+2 | 94 | –15.18 |
| HIVEP zinc finger 2 (HIVEP2) | hsa-miR-532-3p | 76 | –47.97 |
| ubiquitin protein ligase E3 component n-recognin 7 (UBR7) | hsa-miR-139-5p_1ss9GT | 57 | –11.59 |
| ubiquitin protein ligase E3 component n-recognin 7 (UBR7) | hsa-miR-301a-3p | 88 | –27.41 |
| ubiquitin protein ligase E3 component n-recognin 7 (UBR7) | hsa-miR-454-3p_R+2 | 89 | –27.50 |
| indolethylamine N-methyltransferase (INMT) | hsa-miR-486-5p | 55 | –15.57 |
| abhydrolase domain containing 5, lysophosphatidic acid acyltransferase (ABHD5) | hsa-miR-146a-5p_1ss9GT | 94 | –18.01 |
| Gprotein-coupledreceptor class C group 5 member A (GPRC5A) | hsa-miR-1255a | 94 | –51.69 |
| Gprotein-coupledreceptor class C group 5 member A (GPRC5A) | hsa-miR-142-3p_R-1 | 63 | –34.07 |
| Gprotein-coupledreceptor class C group 5 member A (GPRC5A) | hsa-miR-3120-3p_1ss9GT | 55 | –31.85 |
| Gprotein-coupledreceptor class C group 5 member A (GPRC5A) | hsa-miR-532-3p | 93 | –50.65 |
| cytochrome c oxidase assembly homolog COX15 (COX15) | PC-3p-208867_70 | 83 | –17.63 |
To our knowledge, this is the first study to examine the differential expression of circulating microRNAs in HbAS compared with HbAA without concurrent disease states. We identified several differentially expressed circulating plasma microRNAs in HbAS that regulate crucial intracellular processes. These results suggest that the presence of HbAS alone may alter gene expression.
Although there were no comorbidities in this cohort, several of the differentially expressed microRNAs have been associated in the literature with various kidney ischemia-reperfusion injury models (hsa-20a-5p, hsa-486-5p, and hsa-126-3p) and VTE risk (hsa-126-3p and hsa-miR-146a-5p_1ss9GT).11-14 The role of these microRNAs with previously described disease associations in HbAS (pulmonary embolism and CKD)2,3 needs to be further examined. A literature review of the target genes linked to the differentially expressed microRNAs also showed previously reported associations with CKD (CX3CL1 and CD74 genes)15,16 and thrombosis (RALA gene),17 but none were associated with severe sickle cell disease phenotypes.
The differentially expressed microRNAs in the HbAS cohort mapped to several signaling pathways which, in the literature, are associated with chronic tissue inflammation (Axon guidance and p53),18 various cancers (Hippo, phosphatidylinositol 3-kinase/protein kinase B [PI3/Akt], transforming growth factor β, p53, and wingless-related integration site),19 VTE (colorectal cancer, breast cancer, and hepatitis B),20 and CKD (MAPK, phosphatidylinositol 3-kinase/protein kinase B, renin-angiotensin-aldosterone system, transforming growth factor β, wingless-related integration site, Hippo, hypoxia-induced factor 1, ubiquitin-mediated proteolysis, and forkhead box transcription factors).21-24 Understanding how the presence of HbAS may modulate these signaling pathways needs to be clarified.
The results of our study provide interesting data on the differential expression of circulating microRNAs in HbAS. However, the small size of the studied population, the lack of a validation cohort, and the absence of α-thalassemia data are the main limitations of this study. Therefore, these results must be interpreted with caution and replicated using independent cohorts. The number of significantly differentially expressed microRNAs despite the relatively low fold changes highlights the complex biology of HbAS and the need to determine whether these changes are physiologically significant. Future studies involving larger sample sizes, longitudinal data, and compared with participants with specific comorbidities of interest could help increase the specificity of our results to HbAS.
In conclusion, our study demonstrated that HbAS is associated with the significant differential expression of several circulating plasma microRNAs, several of which have previously been associated with kidney injury and VTE in the literature. Further studies are needed to investigate a causal role for these microRNAs in HbAS CKD and VTE associations.
Acknowledgments: Research reported in this publication was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under the Clinical and Translational Science Awards KL2 Program (award number 5KL2TR003981-04) and the University of Texas Southwestern Internal Medicine Pilot and Feasibility grant (K.O.). R.L. is supported by the PKD Foundation Research Grant and National Institute of Diabetes and Digestive and Kidney Diseases R01DK139033. S.H. is supported by the Lina Obeid Chair in Biomedical Sciences at the Renaissance School of Medicine at Stony Brook University.
Contribution: K.O., R.L., S.N., and S.H. proposed the concept of the study; K.O. and S.K. retrieved samples and collected the data; K.O. prepared the draft of the article and revised it under the supervision of R.L., S.N., and S.H.; and all authors analyzed and interpreted the data, and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kabir Olaniran, Department of Internal Medicine, University of Texas Southwestern Medical Center, 5939 Harry Hines Blvd, Dallas, TX 75390; email: kabir.olaniran@utsouthwestern.edu.
References
Author notes
The microRNA data from patients with sickle cell trait and normal hemoglobin phenotype have been deposited in the Gene Expression Omnibus database (accession number GSE285580).
Additional information is available on reasonable request from the corresponding author, Kabir Olaniran (kabir.olaniran@utsouthwestern.edu).