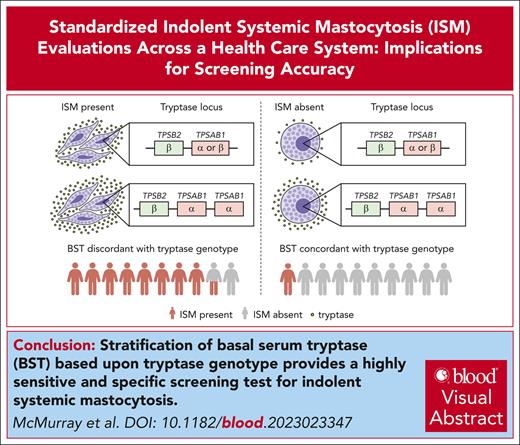
Key Points
An elevated BST based upon genotype is a highly sensitive and specific screening test for indolent SM.
Standardization of screening and diagnostic testing improves SM diagnosis and KIT p.D816V detection rates.
Visual Abstract
Timely diagnosis of systemic mastocytosis (SM) remains challenging because of care heterogeneity. We implemented a standardized approach for SM screening and diagnosis using a novel health care system–wide international screening registry. A retrospective analysis assessed rates of SM, cutaneous mastocytosis (CM), and molecular diagnoses before and 2 years after care standardization. The accuracy of individual and combined SM screening tests, basal serum tryptase (BST) ≥11.5 and ≥20.0 ng/mL, REMA ≥2, monomorphic maculopapular CM (MPCM), and elevated BST based upon tryptase genotype, was analyzed. Tryptase genotyping and high-sensitivity KIT p.D816V testing increased substantially 2 years after care standardization. SM diagnoses doubled from 47 to 94, and KIT p.D816V molecular diagnoses increased from 24 to 79. Mean BST and KIT p.D816V variant allele frequency values were significantly lower in patients diagnosed after standardization. Hereditary-alpha tryptasemia prevalence was increased in SM before care standardization (4/30 [13.3%]) but reflected the general population prevalence 2 years later at (5/76 [6.6%]). Elevated BST based upon genotype and BST ≥11.5 ng/mL had the highest sensitivities at 84.2% and 88.3%, respectively. The presence of monomorphic MPCM, elevated BST based upon tryptase genotype, and the combination of REMA ≥2 with elevated BST based upon tryptase genotype had specificities >90%. BST >20.0 ng/mL had low sensitivity and specificity and was not required to establish any indolent SM (ISM) diagnosis. Care standardization increased SM diagnosis rates, particularly in patients with low BSTs. Stratifying BST based upon genotype had the best overall sensitivity and specificity of any ISM screening test and improved the REMA score specificity.
Introduction
Mastocytosis, as defined by the World Health Organization (WHO) and the International Consensus Classification (ICC),1,2 constitutes a group of rare myeloid neoplasms where the unifying feature is an abnormal accumulation and expansion of neoplastic mast cells (MCs) in 1 or more organ system. The 3 types of mastocytosis include cutaneous mastocytosis (CM), systemic mastocytosis (SM), and MC sarcoma. There are 3 subtypes of CM, including maculopapular CM (MPCM), diffuse CM, and mastocytoma. MPCM can be polymorphic or monomorphic.3 Polymorphic MPCM is nearly always diagnosed in children, whereas monomorphic MPCM is typically diagnosed in adults. SM subtypes include indolent SM (ISM), smoldering SM, aggressive SM, SM with an associated hematologic neoplasm, and mast cell leukemia. The prevalence of SM is estimated to be 1 per 10 000 individuals, and ISM is the most prevalent SM subtype.4-6 The driving mutation in >90% of patients with SM is the somatic variant KIT p.D816V.7,8
SM is important to diagnose early because some subtypes have been linked to reduced survival, disease manifestations reduce quality of life, and new treatment options have become available. Patients with advanced subtypes of SM (aggressive SM, SM-AMN, and mast cell leukemia) and smoldering SM have reduced survival compared with the general population and those with ISM.9 ISM may progress to advanced SM at an estimated rate of 8.4% after 25 years.10 In addition to disease burden affecting survival, up to 56% of patients are at risk of severe or fatal anaphylaxis, especially to hymenoptera envenomation.11,12 Greater than 50% of patients with SM of any subtype are estimated to develop a fragility fracture in their lifetime, and 30% will develop osteoporosis.13-16 The risk of fragility fracture is persistently elevated in patients with SM, despite bisphosphonate therapy.17 Large surveys in both the United States and Europe have shown that cutaneous, gastrointestinal, musculoskeletal, and neurocognitive symptoms in patients with all types of mastocytosis negatively affect quality of life.18-20 Finally, recent treatment advances with selective tyrosine kinase inhibitors for patients with KIT p.D816V+ SM have emphasized the importance of prompt pathological and molecular diagnoses that direct personalized treatment.21-24
There are 3 strategies used to screen for SM. In the first strategy, patients with a clinical or biopsy-proven diagnosis of adult-onset monomorphic MPCM are categorized as high-risk for diagnosis. In fact, 97% of patients with a history of adult-onset MPCM have been reported to have SM.25 By contrast, most children with CM have polymorphic MPCM and are unlikely to be diagnosed with SM.3,26 The second strategy to identify those at high risk of diagnosis of SM involves applying one of a few developed scoring systems that incorporate symptoms experienced during anaphylaxis combined with basal serum tryptase (BST) levels and other factors such as sex or the presence of KIT p.D816V in peripheral blood (PB). In 1 such scoring system known as REMA, proposed by the Spanish Network on Mastocytosis, a score of 2 or higher had a reported specificity of 0.81 and a sensitivity of 0.92 when considering only patients who present with anaphylaxis. Features of anaphylaxis that are high risk for having SM include syncope, presyncope, the absence of urticaria and angioedema, male sex, and BST values >25.0 ng/mL.27 As stated, KIT p.D816V testing of PB has been incorporated into some SM screening strategies.28 However, detectable KIT p.D816V in PB has a reported specificity of 100%, suggesting it may be an ideal diagnostic assay rather than a screening tool.29 The third strategy involves identifying patients with BST values outside the predicted range of their tryptase genotype. Most patients will have a normal tryptase genotype with 1 copy of TPSAB1 on each homologous chromosome, and their BST upper limit of normal is <11.5 ng/mL. Approximately 5% of patients will have an abnormal tryptase genotype known as the hereditary alpha tryptasemia (HαT) genetic trait by having 1 or more extra tandem copies of the TPSAB1 alpha allele. Patients with HαT have BST values with an upper limit of normal that is based upon the number of extra TPSAB1 alpha alleles they have. Patients are thought to be at risk of SM when their BST value is higher than predicted for their tryptase genotype.30,31
Screening for and timely diagnosis of SM are challenging. For instance, it has been reported that 37% of patients with ISM have a subvariant known as bone marrow (BM) mastocytosis and do not have skin involvement, whereas up to 50% of patients or more with SM have no history of anaphylaxis,11,32 and a portion of patients with SM, particularly those with BM mastocytosis, have BST values <11.5 ng/mL.30,33,34 Moreover, tryptase genotyping is recommended by the WHO when an elevated BST is identified in a patient being evaluated for SM to apply the associated minor criterion of >20.0 ng/mL. However, tryptase genotyping only recently became available as a clinical assay. Finally, there is limited sensitivity or specificity data for screening tests, both individually and in combination, to predict the risk of SM. Once screening is completed, there are further challenges in obtaining an accurate diagnosis. Access to mast cell flow cytometry of BM aspirate samples and high-sensitivity quantitative KIT p.D816V assays is somewhat limited, and like tryptase genotyping, the latter has only recently become available. Hematopathology expertise at centers of excellence (COEs) is generally recommended to avoid errors.33,35
On 1 July 2021, we established a health care system–wide, international mastocytosis clinical screening registry for Tricare beneficiaries consisting of military personnel, dependents, foreign nationals, and retirees. The goals of the registry were to enable standardization and centralization of evaluations of patients suspected to have SM across the US Military Health System to improve the quality of care. Here we present a retrospective analysis to assess the impact of the registry on the clinical and molecular diagnoses of patients with SM.
Methods
A retrospective analysis comparing 2 time points, 1 July 2021 and 1 July 2023, was performed on a cohort of Tricare beneficiaries who had a diagnosis of SM or were determined to be at high risk of having SM. The cohort was obtained by querying an international mastocytosis clinical screening registry linked to the Military Health System electronic medical record and claims data accessed from the Military Health System population health portal. Specifically, the registry was queried on 1 July 2021 for individuals with (1) an ICD10 code D47.01 corresponding to CM, (2) ICD10 codes D47.02, C96.21, and C96.29 corresponding to SM subtypes, and (3) any tryptase level (acute or basal) ≥7.0 ng/mL . Patients were determined to be at high risk of an SM diagnosis because of the presence of adult-onset monomorphic MPCM, high-risk anaphylaxis (REMA ≥2), or an elevated BST based upon tryptase genotype.25,27,30 A glossary of terms have been provided in supplemental Material, available on the Blood website. The query searched among current Tricare beneficiaries from 20 large markets, 17 small markets, and many stand-alone clinics dispersed throughout the United States, as well as 1 large market in the Indo-Pacific and 1 large market in Europe.
The SM screening and diagnostic workflows occurred from 1 July 2021 to 1 July 2023. The screening workflow began with chart review to determine whether a patient with CM had monomorphic MPCM, whether a patient with anaphylaxis had a REMA score ≥2, and whether a patient with BST ≥11.5 ng/mL had undergone tryptase genotyping. Patients with a BST value ≥11.5 ng/mL who had not yet been offered tryptase genotyping, were offered testing either in-person or through at-home testing. The diagnostic workflow focused on 4 groups of patients in the registry, including those with: SM, adult-onset monomorphic MPCM, a history of anaphylaxis with REMA score ≥2, an elevated BST based on reported reference ranges for their specific tryptase genotype,31,36 or a combination of these. All patients in these groups were offered a referral to a COE for clinical evaluation. Patients with adult-onset monomorphic MPCM were offered bone marrow biopsy (BMB), KIT p.D816V polymerase chain reaction (PCR) of PB and BM aspirates, and tryptase genotyping. Patients with a documented history of anaphylaxis and an associated REMA score ≥2 were offered BMB, KIT p.D816V PCR of PB and BM aspirate, and tryptase genotyping. Patients with an elevated BST based on genotype were offered a BMB and KIT p.D816V PCR of PB and BM aspirates. Patients already diagnosed with SM were offered high-sensitivity KIT p.D816V PCR testing of PB and tryptase genotyping. SM screening tests and diagnostic quality checks are provided in Table 1.
Standardized SM screening and diagnostic testing implemented 1 July 2021
| . | References . |
|---|---|
| Screening tests | |
| 1. Presence of monomorphic MPCM? (yes or no) | 3,25 |
| 2. Presence of high-risk anaphylaxis (REMA Score ≥ 2)? (yes or no) | 27,54 |
| 3. Elevated BST based upon tryptase genotype? (yes or no) | 30,31 |
| Diagnostic testing quality checks | |
| 1. Was high-sensitivity KIT p.D816V quantitative PCR performed? (yes or no) | 37 |
| 2. Was CD117, tryptase, CD25, CD2 IHC performed? (yes or no) | 55 |
| 3. Was mast cell flow cytometry performed? (yes or no) | 56 |
| 4. Was the BM analyzed at a center of excellence? (yes or no) | 33,35,57 |
| . | References . |
|---|---|
| Screening tests | |
| 1. Presence of monomorphic MPCM? (yes or no) | 3,25 |
| 2. Presence of high-risk anaphylaxis (REMA Score ≥ 2)? (yes or no) | 27,54 |
| 3. Elevated BST based upon tryptase genotype? (yes or no) | 30,31 |
| Diagnostic testing quality checks | |
| 1. Was high-sensitivity KIT p.D816V quantitative PCR performed? (yes or no) | 37 |
| 2. Was CD117, tryptase, CD25, CD2 IHC performed? (yes or no) | 55 |
| 3. Was mast cell flow cytometry performed? (yes or no) | 56 |
| 4. Was the BM analyzed at a center of excellence? (yes or no) | 33,35,57 |
Individual chart review was performed to determine study eligibility (supplemental Figure 1). A patient was included in the study if he or she met any 1 of the 4 criteria: (1) there was a diagnosis of any subtype of SM; (2) there was a diagnosis of adult-onset (age ≥18) monomorphic MPCM; (3) there was at least 1 BST measurement ≥11.5 ng/mL, and tryptase genotyping was performed; or (4) there was a history of anaphylaxis with an associated REMA score ≥2, and either KIT p.D816V was detected in PB or a BMB was performed if KIT p.D816V was not detected in PB. A patient was excluded if he or she met any 1 of the 2 criteria: (1) there was no BST ≥11.5, no history of anaphylaxis with an associated REMA score ≥2, and there was no history of SM or adult-onset monomorphic MPCM at any time point; or (2) they met inclusion criterion #3, had an elevated BST based on their tryptase genotype, and there was no BMB performed, and KIT p.D816V was not detected or tested in PB. This study was approved by the Walter Reed National Military Medical Center Institutional Review Board.
Coded data were collected by reviewing subject charts and recorded into a Microsoft Excel database. Data included age, sex, history of SM, history of monomorphic MPCM, BST values, history of anaphylaxis, tryptase genotype results, KIT p.D816V testing methodology, and results, as well as BMB pathology results. BMB specimens were reviewed by authors who are hematopathologists, I.M. and E.P., and myeloid neoplasm diagnoses were determined using WHO criteria (supplemental Table 1).1 REMA scores were calculated for subjects with a history of anaphylaxis, as previously described.27 Tryptase genotyping was performed at Gene by Gene (Houston, TX), generating TPSAB1 and TPSB2 copy number reports. Previously reported tryptase reference ranges based on tryptase genotypes were used.30 SM screening and diagnostic testing errors were recorded. See supplemental Methods for a description of high-sensitivity KIT p.D816V PCR and tryptase genotyping methodology.
Differences in the distribution of continuous variables BST and KIT p. D816V variant allele frequency (VAF) between groups with SM were analyzed by the Mann-Whitney test. P < .05 was considered statistically significant. Clinical utility of BST, REMA score, tryptase genotyping, and presence of monomorphic MPCM in screening for SM was assessed by calculating sensitivity, specificity, positive likelihood ratio (LR+), negative likelihood ratio (LR–), and Youden indices in patients who underwent BMB or had detectable KIT p.D816V. Definitions of statistical terms have been provided in supplemental Methods. Statistical analyses were performed using IBM SPSS v28, and PRISM 10 (GraphPad Software) was used for graphical illustrations.
Results
Study population
On 1 July 2021, 1420 individuals were identified from a population of 2 752 884 active beneficiaries with a SM diagnosis code, CM diagnosis code, a tryptase ≥7.0 ng/mL, or a combination of these. Three groups were excluded from analysis based on chart data available on 1 July 2021, including: 88 individuals who were miscoded as having mastocytosis or had no records, 690 individuals with BST <11.5 ng/mL and no history of anaphylaxis or with a history of anaphylaxis and REMA <2, and 203 individuals with a CM subtype not linked to SM. Eighty-two individuals on 1 July 2021 had a diagnosis of SM or monomorphic MPCM and were included in the study at the 1 July 2021 time point. On 1 July 2021, 357 individuals did not have a diagnosis of SM or adult-onset monomorphic MPCM but had either (1) a history of anaphylaxis with REMA ≥2 or (2) BST ≥11.5 ng/mL. Clinical evaluations were performed between 1 July 2021 and 1 July 2023 based on clinical screening algorithms (Table 1).
By 1 July 2023, 20 individuals were identified who had a BST <11.4 ng/mL and REMA ≥2, and 337 individuals had a BST ≥11.5 ng/mL. Nine individuals with BST <11.4 ng/mL and REMA ≥2 were excluded because KIT p.D816V was not detected or tested in PB and a BMB was not performed. Eleven individuals with BST <11.4 ng/mL and REMA ≥2 were included because KIT p.D816V was detected in PB or a BMB was performed if KIT p.D816V was not detected in PB. Of those with BST ≥11.5 ng/mL and no mastocytosis diagnosis, 173 patients were excluded because they did not have tryptase genotyping performed. One hundred eight of 173 had no history of anaphylaxis. Of the 65 of 173 with a history of anaphylaxis, 59 (90.8%) had a REMA <2, and 6 (9.2%) had a REMA ≥2. In addition, 8 patients with a normal BST based on their tryptase genotype were excluded because KIT p.D816V was not detected or tested in PB and BMB was not performed. Of those with BST ≥11.5 ng/mL, 156 were included because they had either (1) HαT or (2) a normal tryptase genotype and BMB was performed or KIT p.D816V was detected in PB. The final cohort under study included 249 individuals.
Demographics
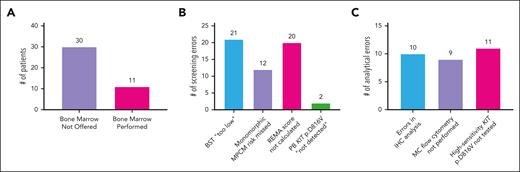
Demographics of the total cohort, the subgroup of patients diagnosed with SM by 1 July 2023, and the subgroup of patients diagnosed with SM by 1 July 2021 are shown in Table 2. Total counts of all myeloid neoplasms are shown in Table 3. Males outnumbered females in the total cohort and SM subgroup. There were 89 patients with myeloid neoplasms and 47 patients with any SM subtype by 1 July 2021 for an SM prevalence of 1 in 58 572 individuals. There were 47 new diagnoses of SM from 1 July 2021 to 1 July 2023. There were 115 patients with myeloid neoplasms and 94 patients diagnosed with any SM subtype by 1 July 2023 for a SM prevalence of 1 in 29 286 individuals. Of the 47 individuals newly diagnosed with SM, 41 had their diagnoses initially missed due to a BMB not being offered or BM analytical errors (Figure 1A). Six of the 47 had no diagnostic delay due to a direct referral to a COE. A BST considered too low for an SM diagnosis was the most frequent screening error (Figure 1B) and a lack of high-sensitivity KIT p.D816V PCR was the most frequent BM analytical error (Figure 1C). In those diagnosed with mastocytosis by 1 July 2021, 3 patients diagnosed with SM had their subtype changed, 22 patients diagnosed with CM were diagnosed with a SM subtype, and 2 patients had other myeloid neoplasms uncovered after standardization of care and expert hematopathology review by authors I.M. and E.P.
Demographics of the study population
| Characteristic . | Total cohort . | SM cohort . | |
|---|---|---|---|
| 1 July 2023 (n = 249) . | 1 July 2023 (n = 94) . | 1 July 2021 (n = 47) . | |
| Age (y), median (IQR) | 46 (37-61) | 47 (37-60) | 47 (38-62) |
| Sex | |||
| Female | 42.2% (105/249) | 37.2% (35/94) | 48.9% (23/47) |
| Male | 57.8% (144/249) | 62.8% (59/94) | 51.1% (24/47) |
| BST levels (ng/mL) | |||
| BST <11.5 (n) | 11.6% (29/249) | 14.9% (14/94) | 4.3% (2/47) |
| BST 11.5 to 14.9 (n) | 23.7% (59/249) | 16.0% (15/94) | 2.1% (1/47) |
| BST 15.0 to 19.9 (n) | 27.7% (69/249) | 17.0% (16/94) | 14.9% (7/47) |
| BST ≥20 (n) | 36.9% (92/249) | 52.1% (49/94) | 78.7% (37/47) |
| HαT tested–positive/total tested (%) | 43.8% (96/219) | 6.6% (5/76)† | 0.0% (0/1)† |
| BST (ng/mL) in HαT+ (mean [95% CI]) | 18.6 (15.3-21.9) | 71.1 (13.6-128.6) | NA |
| BST (ng/mL) in HαT- (mean [95% CI]) | 24.3 (19.5-29.1) | 29.6 (21.9-37.3) | NA |
| History of anaphylaxis (n) | 37.3% (93/249) | 42.6% (40/94) | 29.8% (14/47) |
| REMA score | |||
| REMA ≥ 2 | 60.2% (56/93) | 82.5% (33/40) | 100% (14/14) |
| REMA < 2 | 39.8% (37/93) | 17.5% (7/40) | 0% (0/14) |
| History of monomorphic MPCM (n) | 30.1% (75/249) | 63.8% (60/94) | 85.1% (40/47) |
| KIT p.D816V PB or BM- positive/total (%) | 31.7% (79/249) | 84.0% (79/94)∗ | 51.1% (24/47) |
| Characteristic . | Total cohort . | SM cohort . | |
|---|---|---|---|
| 1 July 2023 (n = 249) . | 1 July 2023 (n = 94) . | 1 July 2021 (n = 47) . | |
| Age (y), median (IQR) | 46 (37-61) | 47 (37-60) | 47 (38-62) |
| Sex | |||
| Female | 42.2% (105/249) | 37.2% (35/94) | 48.9% (23/47) |
| Male | 57.8% (144/249) | 62.8% (59/94) | 51.1% (24/47) |
| BST levels (ng/mL) | |||
| BST <11.5 (n) | 11.6% (29/249) | 14.9% (14/94) | 4.3% (2/47) |
| BST 11.5 to 14.9 (n) | 23.7% (59/249) | 16.0% (15/94) | 2.1% (1/47) |
| BST 15.0 to 19.9 (n) | 27.7% (69/249) | 17.0% (16/94) | 14.9% (7/47) |
| BST ≥20 (n) | 36.9% (92/249) | 52.1% (49/94) | 78.7% (37/47) |
| HαT tested–positive/total tested (%) | 43.8% (96/219) | 6.6% (5/76)† | 0.0% (0/1)† |
| BST (ng/mL) in HαT+ (mean [95% CI]) | 18.6 (15.3-21.9) | 71.1 (13.6-128.6) | NA |
| BST (ng/mL) in HαT- (mean [95% CI]) | 24.3 (19.5-29.1) | 29.6 (21.9-37.3) | NA |
| History of anaphylaxis (n) | 37.3% (93/249) | 42.6% (40/94) | 29.8% (14/47) |
| REMA score | |||
| REMA ≥ 2 | 60.2% (56/93) | 82.5% (33/40) | 100% (14/14) |
| REMA < 2 | 39.8% (37/93) | 17.5% (7/40) | 0% (0/14) |
| History of monomorphic MPCM (n) | 30.1% (75/249) | 63.8% (60/94) | 85.1% (40/47) |
| KIT p.D816V PB or BM- positive/total (%) | 31.7% (79/249) | 84.0% (79/94)∗ | 51.1% (24/47) |
Twenty-nine of the 47 patients diagnosed with SM by 1 July 2021 were tested for HαT between 1 July 2021 and 1 July 2023 and 4 were positive for HαT (4/30). Forty-six of the 47 patients diagnosed with SM between 1 July 2021 and 1 July 2023 were tested for HαT and 1 was positive for HαT.
Nine of 94 patients with SM did not have high-sensitivity PCR testing of KIT p.D816V in PB and/or BM.
Only 1 patient diagnosed with SM by 1 July 2021 was also tested for HαT by 1 July 2021.
Counts of myeloid neoplasms before and after standardization of mastocytosis evaluations
| . | 1 July 2023 (n) . | 1 July 2021 (n) . |
|---|---|---|
| CM | ||
| Adult-onset monomorphic MPCM | 15 | 33 |
| Pediatric-onset monomorphic MPCM | 0 | 1 |
| Pediatric-onset DCM | 2 | 5 |
| SM | ||
| Presumed indolent SM∗ | 7 | 0 |
| Indolent SM† | 79 | 44 |
| Aggressive SM | 3 | 1 |
| SM with an associated myeloid hematologic neoplasm | 3 | 2 |
| Mast cell leukemia† | 1 | 1 |
| Non-SM clonal mast cell disorder | ||
| Monoclonal mast cell activation syndrome | 1 | 0 |
| Other myeloid neoplasms | ||
| Myeloid neoplasm with eosinophilia and FIP1L1-PDGFRα fusion | 2 | 1 |
| Myelodysplastic syndrome | 1 | 1 |
| Chronic myelomonocytic leukemia | 1 | 0 |
| Total | 115 | 89 |
| . | 1 July 2023 (n) . | 1 July 2021 (n) . |
|---|---|---|
| CM | ||
| Adult-onset monomorphic MPCM | 15 | 33 |
| Pediatric-onset monomorphic MPCM | 0 | 1 |
| Pediatric-onset DCM | 2 | 5 |
| SM | ||
| Presumed indolent SM∗ | 7 | 0 |
| Indolent SM† | 79 | 44 |
| Aggressive SM | 3 | 1 |
| SM with an associated myeloid hematologic neoplasm | 3 | 2 |
| Mast cell leukemia† | 1 | 1 |
| Non-SM clonal mast cell disorder | ||
| Monoclonal mast cell activation syndrome | 1 | 0 |
| Other myeloid neoplasms | ||
| Myeloid neoplasm with eosinophilia and FIP1L1-PDGFRα fusion | 2 | 1 |
| Myelodysplastic syndrome | 1 | 1 |
| Chronic myelomonocytic leukemia | 1 | 0 |
| Total | 115 | 89 |
All patients counted on 1 July 2021, continued to be active beneficiaries, and were counted on 1 July 2023. Any counts that decreased from 2021 to 2023 reflect reclassification of the myeloid neoplasm.
Presumed ISM reflects patients who had detectable KIT p.D816V in PB but did not have a BMB by 1 July 2023.
There were 4 patients with a well-differentiated morphologic phenotype by 1 July 2023 including 3 with ISM and 1 with mast cell leukemia.
Reasons for missed SM diagnoses. (A) Use of diagnostic BM in 41 SM cases that were missed by 1 July 2021. (B) Reasons why BM was not offered. (C) BMB errors in cases where BMB was performed. Note: 6 of 47 patients diagnosed with SM after 1 July 2021 did not have their SM diagnoses missed by 1 July 2021 because of being referred directly to a center of excellence. In addition, 14 of 47 patients had IHC errors after 1 July 2021 that required expert hematopathologist evaluation.
Reasons for missed SM diagnoses. (A) Use of diagnostic BM in 41 SM cases that were missed by 1 July 2021. (B) Reasons why BM was not offered. (C) BMB errors in cases where BMB was performed. Note: 6 of 47 patients diagnosed with SM after 1 July 2021 did not have their SM diagnoses missed by 1 July 2021 because of being referred directly to a center of excellence. In addition, 14 of 47 patients had IHC errors after 1 July 2021 that required expert hematopathologist evaluation.
Monomorphic MPCM was identified in 85.1% of patients diagnosed with SM by 1 July 2021, compared with 63.8% in those diagnosed by 1 July 2023. By contrast, a history of anaphylaxis was identified in 29.8% of patients diagnosed with SM by 1 July 2021, compared with 42.6% diagnosed with SM by 1 July 2023. Twelve of the 94 patients (12.8%) with SM had both monomorphic MPCM and a history of high-risk anaphylaxis (REMA ≥ 2) by 1 July 2023. Notably, an absent REMA score calculation in 20 patients with SM represented the second-most common screening error leading to a missed SM diagnosis (Figure 1B). Interestingly, 4 of 94 total patients diagnosed with SM did not have either a history of anaphylaxis with REMA score ≥2 or a history of monomorphic MPCM. These 4 patients underwent diagnostic workup for SM for the following reasons: 2 were incidentally found to have increased MCs on BMB for evaluations of other neoplasms, 1 was found to have an elevated BST during chronic angioedema workup, and 1 was found to have an elevated BST after a history of multiple pathologic fractures.
BST values and tryptase genotyping
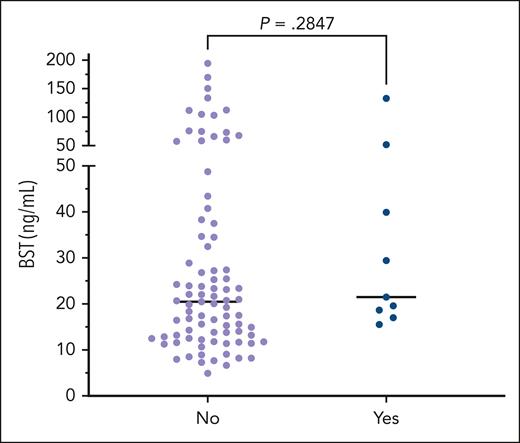
Median BST (interquartile range) was 14.9 ng/mL (11.7-21.6) in the total cohort. Median BST was significantly higher in patients with SM diagnosed by 1 July 2021 (27.2 ng/mL [20.7-66.9]) than in those diagnosed with SM by 1 July 2023 (20.5 ng/mL [13.3-36.7]); P = .002. BST values ≥20.0 ng/mL were overrepresented, and BST values <15 ng/mL were underrepresented in patients diagnosed with SM by 1 July 2021, relative to those diagnosed by 1 July 2023 (Table 2). Mean BST values were significantly higher in the 47 patients diagnosed with SM by 1 July 2021 (50.2 ng/mL) compared with the 47 diagnosed after 1 July 2021 (19.3 ng/mL); P < .0001 (Figure 2A). None of the 49 patients with SM who had BST >20.0 ng/mL required the BST >20.0 ng/mL to be diagnosed with SM because other criteria were met (supplemental Table 1 for WHO criteria). Nine patients with SM had a history of ≥1 pathologic fracture (mean, 2.3). Of those 9 patients, 4 had osteopenia, 4 had osteoporosis, and 1 did not have a DEXA scan for review. Mean BST values were not significantly different between patients with SM with and without a history of pathologic fracture (Figure 3).
SM disease burden biomarkers before and after care standardization. BST (A) and KIT p.D816V VAF (B) distributions on continuous scatter plots comparing SM cases diagnosed before 1 July 2021 to those diagnosed after 1 July 2021. In panels A and B, blue dots correspond to SM cases diagnosed by 1 July 2021, and purple dots correspond to SM cases diagnosed after 1 July 2021. In panel B, red dots represent SM cases diagnosed by 1 July 2021 where KIT p.D816V VAF values were tested after 1 July 2021. Each patient’s highest lifetime KIT p.D816V VAF value of either PB or BM aspirate was used in panel B. PB and BM aspirate were also analyzed separately (see supplemental Figure 2). Mann-Whitney test was used (P value) to compare means of the 2 groups.
SM disease burden biomarkers before and after care standardization. BST (A) and KIT p.D816V VAF (B) distributions on continuous scatter plots comparing SM cases diagnosed before 1 July 2021 to those diagnosed after 1 July 2021. In panels A and B, blue dots correspond to SM cases diagnosed by 1 July 2021, and purple dots correspond to SM cases diagnosed after 1 July 2021. In panel B, red dots represent SM cases diagnosed by 1 July 2021 where KIT p.D816V VAF values were tested after 1 July 2021. Each patient’s highest lifetime KIT p.D816V VAF value of either PB or BM aspirate was used in panel B. PB and BM aspirate were also analyzed separately (see supplemental Figure 2). Mann-Whitney test was used (P value) to compare means of the 2 groups.
BST distributions on a continuous scatterplot comparing patients with SM with and without a history of pathologic fractures. The y-axis corresponds to BST values. Patients with SM with a history of pathologic fracture are grouped in the “Yes” column and those patients with SM without a history of pathologic fracture are grouped in the “No” column.
BST distributions on a continuous scatterplot comparing patients with SM with and without a history of pathologic fractures. The y-axis corresponds to BST values. Patients with SM with a history of pathologic fracture are grouped in the “Yes” column and those patients with SM without a history of pathologic fracture are grouped in the “No” column.
Most of the tryptase genotyping was performed after 1 July 2021 (Figure 4A). Tryptase genotyping was completed in only 5 patients by 1 July 2021, including in 1 patient with SM. Tryptase genotyping was completed in 214 patients between 1 July 2021 and 1 July 2023, including 75 patients with SM. HαT was identified in 43.8% (n = 96) of all 219 patients tested (Figure 4B). Individuals diagnosed with HαT had BST and tryptase genotyping tested for a clinical history of urticaria 35.4% (n = 34), anaphylaxis 21.9% (n = 21), family history of HαT 9.4% (n = 9), eosinophilia 7.3% (n = 7), and other problems 26.0% (n = 25). In the subgroup of 47 patients diagnosed with SM by 1 July 2021, 13.3% (n = 4) of the 30 patients tested were subsequently found to have HαT. In the 47 patients diagnosed with SM between 1 July 2021 and 1 July 2023, only 1 additional patient was diagnosed with HαT, for a final prevalence of 6.6% (n = 5) in the 76 patients with SM tested. One of the 5 patients with SM and HαT had BST values within reported reference ranges for their tryptase genotype. Eighteen of 94 patients with SM did not have tryptase genotyping, and 6 of 18 had BST >35.0 ng/mL. Fourteen patients with monomorphic MPCM did not have tryptase genotyping, and 1 of 14 had BST >35.0 ng/mL.
Summary of molecular testing. (A) Comparison of molecular testing performed before and after 1 July 2021. Patients were counted both before and after 1 July 2021 if high sensitivity PCR testing was performed in both. For PB testing, 4 patients had PCR testing both before and after 1 July 2021. Of those with PB PCR testing, 159 had 1 test and 15 had 2 or more PCR tests during either the before and/or after 1 July 2021 time points. For BM testing, 2 patients had PCR testing both before and after 1July 2021. Of those with BM PCR testing, 71 had 1 testing method and 10 had 2 or more testing methods performed (See supplemental Methods). (B) Distribution of tryptase genotypes in both the total cohort excluding patients with SM and in the subgroup of patients with SM. Four patients in the total cohort excluding patients with SM were tryptase genotyped before 1 July 2021 and 1 patient in the SM subgroup was tryptase genotyped before 1 July 2021. (C) Comparison of KIT p.D816V positive results by testing modalities performed before and after 1 July 2021. Fifty-nine patients had only 1 positive testing method and 20 patients had 2 or more positive testing methods by either PB and/or BM during either before and/or after 1 July 2021 time points.
Summary of molecular testing. (A) Comparison of molecular testing performed before and after 1 July 2021. Patients were counted both before and after 1 July 2021 if high sensitivity PCR testing was performed in both. For PB testing, 4 patients had PCR testing both before and after 1 July 2021. Of those with PB PCR testing, 159 had 1 test and 15 had 2 or more PCR tests during either the before and/or after 1 July 2021 time points. For BM testing, 2 patients had PCR testing both before and after 1July 2021. Of those with BM PCR testing, 71 had 1 testing method and 10 had 2 or more testing methods performed (See supplemental Methods). (B) Distribution of tryptase genotypes in both the total cohort excluding patients with SM and in the subgroup of patients with SM. Four patients in the total cohort excluding patients with SM were tryptase genotyped before 1 July 2021 and 1 patient in the SM subgroup was tryptase genotyped before 1 July 2021. (C) Comparison of KIT p.D816V positive results by testing modalities performed before and after 1 July 2021. Fifty-nine patients had only 1 positive testing method and 20 patients had 2 or more positive testing methods by either PB and/or BM during either before and/or after 1 July 2021 time points.
Detection of KIT p.D816V
Patients diagnosed with SM by 1 July 2021 more often had low-sensitivity KIT Sanger sequencing as well as qualitative rather than quantitative KIT p.D816V PCR testing compared with patients diagnosed with SM after 1 July 2021 (Figure 4A). Only 23 of 47 patients (48.9%) with SM had 1 or more high-sensitivity KIT p.D816V PCR tests performed on PB, BM, or both by 1 July 2021. Sixty-five more patients with SM, for a total of 88 of 94 (93.6%), had 1 or more high-sensitivity KIT p.D816V PCR tests performed on PB, BM, or both by 1 July 2023. A total of 51.1% (n = 24) patients with SM had KIT p.D816V detected by PCR or next generation sequencing (NGS) of PB, BM, or both by 1 July 2021, compared with 84.0% (n = 79) by 1 July 2023. One of the 47 patients (2.1%) with SM had a positive NGS test, 23 of the 47 patients (48.9%) with SM had a positive PCR test, and 0 patients with SM had both positive NGS and PCR tests by 1 July 2021. From 1 July 2021 to 1 July 2023, no additional patients had a positive NGS test only (n = 1), 52 additional patients had a positive PCR test only (n = 75), and 3 additional patients had both positive NGS and PCR tests (n = 3) (Figure 4C). When considering only patients with an advanced SM subtype, 2 of 3 patients with SM with an associated hematologic neoplasm had a positive PCR test, and 3 of 3 had a negative NGS test, whereas 3 of 3 patients with aggressive SM had a positive PCR test and 3 of 3 had a negative NGS test. Patients diagnosed with SM by 1 July 2021 had a higher mean KIT p.D816V VAF (2.2% VAF) of the PB and/or BM compared with patients diagnosed with SM after 1 July 2021 (0.3% VAF); P = .0124 (Figure 2B; supplemental Figure 2).
SM screening test accuracy
The sensitivity, specificity, Youden Index, LR+, and LR– of the following 6 screening tests were analyzed for utility in accurately predicting SM: BST ≥11.5 ng/mL, BST ≥20.0 ng/mL, BST elevated based upon genotype, REMA score ≥2, REMA score ≥2 combined with elevated BST based upon genotype, and presence of monomorphic MPCM (Table 4; supplemental Table 3). Patients were included in the analysis only if they underwent BMB or had detectable KIT p.D816V in PB. Only 2 tests had sensitivities over 80% and were BST ≥11.5 ng/mL and BST elevated based on genotype. BST elevated based upon genotype had the most favorable LR–. Three tests demonstrated specificities >90% and positive LRs >8 including presence of monomorphic MPCM, REMA score ≥2 combined with elevated BST based upon genotype, and elevated BST based upon genotype. Notably, the REMA score ≥2 demonstrated a reduced specificity compared with REMA score ≥2 combined with elevated BST based upon genotype, whereas the sensitivity between the 2 was similar. By contrast, BST ≥20.0 ng/mL was the fourth-most sensitive screening test with a sensitivity of 59.6% and the fifth-most specific screening test with a specificity of 76.3%. Of the screening tests analyzed, the Youden’s index was highest with an elevated BST based upon genotype and lowest with a REMA score ≥2. A receiver operating characteristic curve for BST was generated, which demonstrated the optimal cut-off for BST of >20.4 ng/mL based on the highest Youden’s index (0.35) with a sensitivity of 58.5% and a specificity of 76.2% (supplemental Figure 3).
Comparison of sensitivities, specificities, Youden’s indices, and likelihood ratios among SM screening tests
| . | Sensitivity % . | Specificity % . | Youden’s index . | LR+ . | LR– . |
|---|---|---|---|---|---|
| BST ≥11.5 ng/mL (n = 136) | 88.3 | 23.8 | 0.12 | 1.16 (0.96-1.39) | 0.49 (0.23-1.07) |
| BST ≥20.0 ng/mL (n = 136) | 59.6 | 71.4 | 0.31 | 2.09 (1.26-3.46) | 0.57 (0.41-0.77) |
| BST elevated based upon genotype (n = 117) | 84.2 | 90.2 | 0.74 | 8.63 (3.39-22.00) | 0.17 (0.10-0.30) |
| REMA ≥2 (n = 136) | 34.0 | 73.8 | 0.08 | 1.30 (0.73-2.32) | 0.89 (0.71-1.13) |
| REMA ≥2 and BST elevated based upon genotype (n = 117) | 30.3 | 97.6 | 0.28 | 12.00 (1.74-89.00) | 0.71 (0.61-0.84) |
| Monomorphic MPCM (n = 136) | 63.8 | 97.6 | 0.61 | 27.00 (3.84-187.00) | 0.37 (0.28-0.49) |
| . | Sensitivity % . | Specificity % . | Youden’s index . | LR+ . | LR– . |
|---|---|---|---|---|---|
| BST ≥11.5 ng/mL (n = 136) | 88.3 | 23.8 | 0.12 | 1.16 (0.96-1.39) | 0.49 (0.23-1.07) |
| BST ≥20.0 ng/mL (n = 136) | 59.6 | 71.4 | 0.31 | 2.09 (1.26-3.46) | 0.57 (0.41-0.77) |
| BST elevated based upon genotype (n = 117) | 84.2 | 90.2 | 0.74 | 8.63 (3.39-22.00) | 0.17 (0.10-0.30) |
| REMA ≥2 (n = 136) | 34.0 | 73.8 | 0.08 | 1.30 (0.73-2.32) | 0.89 (0.71-1.13) |
| REMA ≥2 and BST elevated based upon genotype (n = 117) | 30.3 | 97.6 | 0.28 | 12.00 (1.74-89.00) | 0.71 (0.61-0.84) |
| Monomorphic MPCM (n = 136) | 63.8 | 97.6 | 0.61 | 27.00 (3.84-187.00) | 0.37 (0.28-0.49) |
Patients were included in this analysis only if they had either (1) a BMB or (2) detectable KIT p.D816V in PB. For the LR+ and LR–, the numbers in brackets represent the 95% confidence intervals. Youden’s index = (sensitivity + specificity) – 1.
Discussion
Evaluations for SM are challenging for a number of reasons, including: (1) SM is a rare disease that the average subspecialist encounters infrequently; (2) the clinical presentation is variable, including monomorphic MPCM, anaphylaxis with REMA ≥2, both, or neither; (3) high-complexity molecular assays, including tryptase genotyping and high-sensitivity KIT p.D816V PCR, are not widely available; (4) validation of rarely used immunohistochemical staining, including tryptase and CD25, is difficult in centers with low volume; and (5) hematopathology expertise is not uniform. These problems partly explain why there is a median diagnostic delay of 9 years for patients with ISM and 7 years for patients with any SM subtype.37,38 Here, we showed that generating a clinical registry and standardizing and centralizing evaluations for SM at COEs increased the frequency of timely and correct clinical and molecular diagnoses of SM.
To our knowledge, we are the first health care system to create such a SM screening and clinical care registry using a systematic approach across a large geographic distance. The patients we analyzed were preselected for contact and referral to a COE if they had a history of adult-onset monomorphic MPCM, anaphylaxis with an associated REMA score ≥2, or an elevated BST based upon tryptase genotype. This approach doubled the prevalence of SM over 2 years and correctly diagnosed 41 of 47 patients (87.2%) with SM who had previously been missed. Of those missed previously, 32 of 41 (78.0%) had BST values <20.0 ng/mL. Similarly, KIT p.D816V VAFs were significantly lower (mean, 0.3% VAF) in those diagnosed with SM after 1 July 2021 (Figure 2B) compared with patients diagnosed by 1 July 2021 (mean, 2.2% VAF). This difference is likely due to the application of high-sensitivity KIT p.D816V quantitative PCR in patients with a lower disease burden. Overall, 32 of 60 patients (53.3%) with SM with detectable KIT p.D816V by a quantitative assay (PCR or NGS) had a VAF <0.1%. This demonstrates the value of high-sensitivity KIT p.D816V PCR testing in both the PB and BM aspirates for the diagnosis of SM. We also showed that immunohistochemistry (IHC) errors are common and occurred in 24 of 94 patients (25%) diagnosed with SM. IHC problems included absent CD25 and tryptase IHC test ordering, CD25 IHC methodology errors, and hematopathologist interpretive errors when assessing major and minor criteria. Importantly, our results demonstrate that molecular testing must be reviewed closely in all patients with SM, but especially in those who are reported to be KIT p.D816V negative, because our data show that this often occurs when only low-sensitivity KIT p.D816V testing, such as Sanger sequencing or NGS, is used.
There are several possible reasons why patients with lower BST and KIT p.D816V VAF values were diagnosed with SM after care standardization. First, patients with SM were frequently not referred for BMB based on the assumption that their BST was too low to have SM. In fact, 12 of 47 patients (25.5%) diagnosed with SM after 1 July 2021 had BST <11.5 ng/mL. Further, the median BST of the 76 patients with SM who underwent tryptase genotyping in our cohort was 18.9 ng/mL and this value was lower than median BST values in prior studies (Table 5).9,39,40 The reported prevalence of HαT in patients with SM has ranged from 12.2% to 21.3%.31,39,41-45 By contrast, we found that the prevalence of HαT in patients with SM in our cohort was no different than the prevalence of HαT in the general population. Our data showed that there can be bias against evaluating patients with low BST values for SM. It is also likely that patients with lower BST and VAF of KIT p.D816V are missed because of limited access to or application of high-complexity molecular testing that is not routinely available outside of large academic centers. Our clinical registry allowed us to offer tryptase genotyping to patients at home as well as high-sensitivity KIT p.D816V PCR remotely in affiliated clinical laboratories in a geographically dispersed population.
Comparison of SM cohorts and prevalence of HαT in published studies
| Study . | Sample (n) . | Population type . | Age, y, median (range) . | Sex (M) . | BST, median (IQR) . | BST (min-max) . | HαT prevalence (%) . |
|---|---|---|---|---|---|---|---|
| Current study | 76 | Non-AdvSM/AdvSM | 49 (35-59.5)∗ | 65.8% | 18.9 (13.1-34.5) | 6.7-171.8 | 6.6 |
| Lyons et al39 | 82 | Non-AdvSM | 47 (21-64)∗ | 45.1% | 38.8 (28.6-67.5) | 8.1-487 | 12.2 |
| Sordi et al37 | 444 | CM/MMAS/non-AdvSM/AdvSM | 51 (40-64)∗ | 55.2% | 25.5 (14.6-52.8) | NA | 13.3 |
| Polivka et al42 | 583 | CM/MMAS/non-AdvSM/AdvSM/MCS | 38 (0-85) | 46.0% | 62.6 (NA) | 1.4-1240 | 13.6 |
| Greiner et al (main study)40 | 180 | CM/non-AdvSM/AdvSM | 48 (11-91) | 45.6% | 36.6 (NA) | 1.8-1617 | 17.2 |
| Chovanec et al30 | 119 | DCM/MMAS/non-AdvSM | NA | NA | NA | NA | 17.7 |
| Gonzalez-de-Olano et al43 | 464 | CM/non-AdvSM/AdvSM | NA (17-83) | 57.5% | 33.6 (NA) | 2.9-1360 | 18.1 |
| Chollet et al41 | 49 | CM/non-AdvSM/AdvSM | NA (23-75) | 22.4% | NA | NA | 18.4 |
| Greiner et al (validation cohort)40 | 61 | Non-AdvSM | 49 (23-71) | 42.6% | 39.0 (NA) | 6.6-163.0 | 21.3 |
| Study . | Sample (n) . | Population type . | Age, y, median (range) . | Sex (M) . | BST, median (IQR) . | BST (min-max) . | HαT prevalence (%) . |
|---|---|---|---|---|---|---|---|
| Current study | 76 | Non-AdvSM/AdvSM | 49 (35-59.5)∗ | 65.8% | 18.9 (13.1-34.5) | 6.7-171.8 | 6.6 |
| Lyons et al39 | 82 | Non-AdvSM | 47 (21-64)∗ | 45.1% | 38.8 (28.6-67.5) | 8.1-487 | 12.2 |
| Sordi et al37 | 444 | CM/MMAS/non-AdvSM/AdvSM | 51 (40-64)∗ | 55.2% | 25.5 (14.6-52.8) | NA | 13.3 |
| Polivka et al42 | 583 | CM/MMAS/non-AdvSM/AdvSM/MCS | 38 (0-85) | 46.0% | 62.6 (NA) | 1.4-1240 | 13.6 |
| Greiner et al (main study)40 | 180 | CM/non-AdvSM/AdvSM | 48 (11-91) | 45.6% | 36.6 (NA) | 1.8-1617 | 17.2 |
| Chovanec et al30 | 119 | DCM/MMAS/non-AdvSM | NA | NA | NA | NA | 17.7 |
| Gonzalez-de-Olano et al43 | 464 | CM/non-AdvSM/AdvSM | NA (17-83) | 57.5% | 33.6 (NA) | 2.9-1360 | 18.1 |
| Chollet et al41 | 49 | CM/non-AdvSM/AdvSM | NA (23-75) | 22.4% | NA | NA | 18.4 |
| Greiner et al (validation cohort)40 | 61 | Non-AdvSM | 49 (23-71) | 42.6% | 39.0 (NA) | 6.6-163.0 | 21.3 |
AdvSM, advanced SM; IQR, interquartile range; M, male; max, maximum; MCS, mast cell sarcoma; min, minimum; NA, not available.
Range refers to IQR, 25th and 75th percentile, if available. Otherwise, min and max are included in range. AdvSM subtypes include aggressive SM, SM with an associated hematologic neoplasm, and mast cell leukemia. Nonadvanced SM (non-AdvSM) subtypes include BM mastocytosis, presumed ISM, ISM, WDSM, and SSM.
We previously reported that BST ≥11.5 ng/mL in the setting of a normal tryptase genotype is linked to SM30 and 1 additional study confirmed these findings.31 The present study is the first to validate that an elevated BST based on genotype is an individually sensitive and specific screening test for SM. An elevated BST based on genotype had the second highest sensitivity and second highest specificity of all individual SM screening tests analyzed. Notably, elevated BST based on genotype had a higher sensitivity and specificity than REMA ≥2 as well as BST >20.0 ng/mL. The combination of REMA ≥2 with elevated BST based on genotype had improved specificity with similar sensitivity compared with REMA ≥2 alone, strongly suggesting the combination of the 2 tests should routinely be used. Specificity did not improve when combining MPCM with elevated BST based upon genotype (data not shown). These results have led us to generate an ISM screening strategy using individual and combination screening tests to improve the detection of ISM (Figure 5). A full description of which screening tests should be performed on which patients has been provided in Figure 5. Most of the patients in our cohort had ISM. Therefore, the screening strategy we developed is relevant for ISM, and its value in screening for other subtypes is not known. It is important to note that the screening strategy we present also has the potential to reduce the number of unnecessary BMBs for patients with HαT, who do not have risk factors for ISM and whose BST values are concordant with their tryptase genotype.
ISM screening algorithm. The algorithm was designed based on individual as well as combined test accuracy in predicting ISM (see supplemental Table 3). In the SM subgroup of patients we presented, 4 of 94 had neither monomorphic MPCM or high-risk anaphylaxis (REMA ≥2). Patients with a history of CM should screened by physical examination for the monomorphic MPCM subtype. All patients with the monomorphic MPCM subtype should be offered a BMB and high-sensitivity KIT p.D816V PCR testing. Patients with a history of anaphylaxis should be screened using the REMA score which also requires measurement of BST. Those with anaphylaxis with an associated REMA score ≥2 with BST values ≥11.5 ng/mL should also be screened with tryptase genotyping. Those with REMA score ≥2 and an elevated BST based upon genotype should be offered a BMB and high-sensitivity KIT p.D816V PCR testing. Patients with a REMA score ≥2 and a BST <11.5 ng/mL should be offered a BMB and high-sensitivity KIT p.D816V PCR testing on a case-by-case basis depending on factors such as patient age, trigger of anaphylaxis (eg, flying hymenoptera), and number of episodes of anaphylaxis with a REMA score ≥2. ∗It should also be noted that patients with ISM may present with other risk factors such as pathologic fractures, flushing, and mast cell hyperplasia in extracutaneous tissues rather than monomorphic MPCM or anaphylaxis. Patients with these risk factors without a clearly-defined cause, should also be screened with tryptase genotyping if the BST is ≥11.5 ng/mL. The accuracy of this screening approach in patients with SM subtypes other than ISM is not known.
ISM screening algorithm. The algorithm was designed based on individual as well as combined test accuracy in predicting ISM (see supplemental Table 3). In the SM subgroup of patients we presented, 4 of 94 had neither monomorphic MPCM or high-risk anaphylaxis (REMA ≥2). Patients with a history of CM should screened by physical examination for the monomorphic MPCM subtype. All patients with the monomorphic MPCM subtype should be offered a BMB and high-sensitivity KIT p.D816V PCR testing. Patients with a history of anaphylaxis should be screened using the REMA score which also requires measurement of BST. Those with anaphylaxis with an associated REMA score ≥2 with BST values ≥11.5 ng/mL should also be screened with tryptase genotyping. Those with REMA score ≥2 and an elevated BST based upon genotype should be offered a BMB and high-sensitivity KIT p.D816V PCR testing. Patients with a REMA score ≥2 and a BST <11.5 ng/mL should be offered a BMB and high-sensitivity KIT p.D816V PCR testing on a case-by-case basis depending on factors such as patient age, trigger of anaphylaxis (eg, flying hymenoptera), and number of episodes of anaphylaxis with a REMA score ≥2. ∗It should also be noted that patients with ISM may present with other risk factors such as pathologic fractures, flushing, and mast cell hyperplasia in extracutaneous tissues rather than monomorphic MPCM or anaphylaxis. Patients with these risk factors without a clearly-defined cause, should also be screened with tryptase genotyping if the BST is ≥11.5 ng/mL. The accuracy of this screening approach in patients with SM subtypes other than ISM is not known.
The current WHO minor criteria for the diagnosis of SM include clonal markers such as CD25 expression on MCs and the KIT p.D816V mutation. BST >20 ng/mL was added as a WHO minor criterion in 200146 before the addition of CD25.29 BST elevations were first reported in SM in 198747 and other myeloid neoplasms later.48-51 However, BST elevations are most frequently seen in HαT.30,52 We recently reported that BST levels did not contribute to the diagnosis of SM in a small cohort when all other diagnostic testing was adequate.33 In the present study with a larger cohort, none of the patients with SM required BST >20.0 ng/mL to be diagnosed with SM because they met other WHO criteria. Other authors have also shown that using BST >20.0 ng/mL may lead to patients with lower BST values being missed.30,31,53 Even though BST >20.0 ng/mL is still a SM criterion by WHO and ICC, it has a number of limitations, does not apply when an associated myeloid hematologic neoplasm is found, has to be corrected in the case of HαT, and may not be fulfilled in a certain proportion of patients with true SM. Given these limitations, the maintenance of this criterion in the WHO and ICC criteria could be a matter of debate in the future.
There are several limitations to this study, including its retrospective design, the fact that some patients were not able to undergo a complete evaluation, and the fact that our SM cohort was comprised predominantly of patients with ISM. For instance, some patients with SM and monomorphic MPCM did not undergo tryptase genotyping, and this may have affected the prevalence calculation of HαT in SM. Nonetheless, the unique design of the clinical screening registry in proactively and systematically offering referrals based upon documented diagnoses and laboratory values to COEs in a geographically dispersed patient population may have minimized referral bias compared with other registry studies that rely on community physician referrals. It is also important to note that tryptase genotyping is only available at 1 laboratory vendor in the United States, and this might limit the ability of clinicians to implement the ISM screening approach we describe. To overcome this challenge, we suggest considering tryptase genotyping using at-home testing through partnerships with COEs for patients living outside of academic centers until tryptase genotyping becomes more widely available. Finally, our screening test accuracy findings are most applicable to those with nonadvanced SM because only a few patients we presented had an advanced SM subtype.
In summary, we have shown that a unique clinical SM screening and care registry linked to COEs can improve SM pathological and KIT p.D816V molecular diagnosis rates over regular referral pathways. We have also elucidated reasons for missed diagnoses of SM, including bias against referring patients with low BST for diagnostic testing, failure to interpret the contribution of anaphylaxis and monomorphic MPCM to pretest probability, a lack of high-sensitivity KIT p.D816V testing, and the need for hematopathology expertise. Finally, we have shown that 2 individual screening tests (monomorphic MPCM and elevated BST based upon genotype) and 1 combination of 2 screening tests (REMA ≥ 2 with elevated BST based upon genotype) have utility in ISM screening.
Acknowledgments
The authors thank Sofia C. Echelmeyer of the Uniformed Services University for her assistance with the visual abstract illustration.
This research was funded in whole or in part by the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, Uniformed Services University, or Department of Defense, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
Authorship
Contribution: J.C.M., N.A.B., and I.M. designed the study; I.M. and E.M.P. evaluated bone marrow biopsy tissue; J.C.M., N.A.B., B.J.S., C.S.P., J.A.B., A.E.S., J.S.A., R.C.C., L.M.P., K.E.A., A.M.W., E.M.P., J.J.L., D.D.M., I.M., C.-T.J.K., and X.S. generated the study data; J.C.M. performed data analysis; and J.C.M., N.A.B., and B.J.S. wrote the manuscript with input from all coauthors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nathan A. Boggs, Uniformed Services University, 4301 Jones Bridge Rd, Bethesda, MD 20814; email: nathan.boggs@usuhs.edu.
References
Author notes
Data are available on request from the corresponding author, Nathan A. Boggs (nathan.boggs@usuhs.edu).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal