Key Points
Time-limited acalabrutinib, venetoclax, and obinutuzumab induced deep and ongoing remission in patients with relapsed/refractory CLL.
The addition of ctDNA-based analyses to conventional FCM appears to improve early detection of molecular and clinical relapses.
Visual Abstract
The phase 2 CLL2-BAAG trial tested the measurable residual disease (MRD)–guided triple combination of acalabrutinib, venetoclax, and obinutuzumab after optional bendamustine debulking in 45 patients with relapsed/refractory chronic lymphocytic leukemia (CLL). MRD was measured by flow cytometry (FCM; undetectable MRD <10–4) in peripheral blood (PB) and circulating tumor DNA (ctDNA) using digital droplet polymerase chain reaction of variable-diversity-joining (VDJ) rearrangements and CLL-related mutations in plasma. The median number of previous treatments was 1 (range, 1-4); 18 patients (40%) had received a Bruton tyrosine kinase inhibitor (BTKi) and/or venetoclax before inclusion, 14 of 44 (31.8%) had TP53 aberrations, and 34 (75.6%) had unmutated immunoglobulin heavy-chain variable region genes. With a median observation time of 36.3 months and all patients off-treatment for a median of 21.9 months, uMRD <10–4 in PB was achieved in 42 of the 45 patients (93.3%) at any time point, including 17 of 18 (94.4%) previously exposed to venetoclax/BTKi and 13 of 14 (92.9%) with TP53 aberrations. The estimated 3-year progression-free and overall survival rates were 85.0% and 93.8%, respectively. Overall, 585 paired FCM/ctDNA samples were analyzed and 18 MRD recurrences (5 with and 13 without clinical progression) occurred after the end of treatment. Twelve samples were first detected by ctDNA, 3 by FCM, and 3 synchronously. In conclusion, time-limited MRD-guided acalabrutinib, venetoclax, and obinutuzumab achieved deep remissions in almost all patients with relapsed/refractory CLL. The addition of ctDNA-based analyses to FCM MRD assessment seems to improve early detection of relapses. This trial was registered at www.clinicaltrials.gov as #NCT03787264.
Introduction
The landscape of chronic lymphocytic leukemia (CLL) treatment has undergone a significant transformation in recent years.1 The emergence of targeted agents, such as Bruton tyrosine kinase (BTK) inhibitors and B-cell lymphoma 2 (Bcl-2) inhibitors, has led to substantial improvements in patient outcomes, spanning all patient and risk groups.2-15 In recent years, triple combinations using a BTK inhibitor (BTKi), venetoclax, and an anti-CD20 antibody have been tested in various settings and patient groups.2,16-20 In the phase 3 GAIA/CLL13 trial, a first-line combination of venetoclax, ibrutinib, and obinutuzumab achieved an undetectable measurable residual disease (uMRD) rate in peripheral blood (PB) of 92.2% posttreatment translating into a high estimated 3-year progression-free survival (PFS) rate of 90.5%.2 Several phase 2 investigations have corroborated these findings in the first-line setting using different BTKi: a measurable residual disease (MRD)-guided therapy with acalabrutinib, venetoclax, and obinutuzumab led to a PB uMRD rate of 86% after 15 months of treatment and in the BOVen trial, zanubrutinib, venetoclax, and obinutuzumab led to a similar PB uMRD rate (96%).19,21 Among patients with TP53 aberrations, MRD-guided first-line treatment with ibrutinib, venetoclax, and obinutuzumab achieved a PB uMRD rate of 78% at 15 months, which translated into a 3-year PFS rate of 79.9%.22 However, in a relapsed/refractory setting, particularly in patients with relapsed disease after BTKi and/or venetoclax, the available evidence is limited, necessitating further exploration of optimal therapeutic approaches. The CLL2-BAAG trial tested a combination of acalabrutinib, venetoclax, and obinutuzumab, after optional debulking with bendamustine in patients with relapsed/refractory CLL.18 In its primary end point analysis, the trial demonstrated deep remissions with a uMRD rate of 75.6% in PB upon final restaging after 6 months of the triplet. In this analysis, the long-term efficacy of MRD-guided combination therapy was evaluated in a population characterized by a high proportion of patients with high-risk genetics and/or previous exposure to venetoclax and/or BTKi. Furthermore, the feasibility and performance of an MRD-guided approach were evaluated to customize the treatment duration to the needs of differently pretreated patients with different risk profiles.
Recently, the use of circulating tumor DNA (ctDNA) for disease monitoring, detection, and prognostication has gained relevance, particularly in the field of lymphoma.23-28 An initial prospective evaluation of ctDNA in CLL demonstrated the applicability of complementary ctDNA-based MRD monitoring and highlighted several potential advantages compared with conventional flow cytometry (FCM).29 Notably, ctDNA appeared to provide a more accurate reflection of the disease burden across various compartments and seemed to more sensitively detect residual disease in patients with residual lymph nodes. This analysis set out to evaluate whether ctDNA also has value in the early detection of molecular and clinical relapses.
Methods
Patient population
In the multicenter, investigator-initiated phase 2 CLL2-BAAG study the combination of obinutuzumab, acalabrutinib, and venetoclax after optional debulking with bendamustine was assessed in patients with relapsed/refractory CLL. During screening, next generation sequencing (NGS)-based testing for mutations in BTK and PLCG2 was performed in patients pre-exposed to BTKi, and patients with known resistance-conferring mutations were excluded from participation. Obinutuzumab was started in the first cycle (days 1, 8, and 15), in cycle 2 acalabrutinib was added, and in cycle 3 venetoclax ramp up (over 5 weeks up to 400 mg) was initiated. All 3 drugs were administered according to the established schedules with daily acalabrutinib, venetoclax, and obinutuzumab once every 4 weeks during induction and every 12 weeks during maintenance (Figure 1A).18 Induction treatment was administered for a total of 8 cycles (ie, 6 cycles of the triple combination) until final restaging before the patients entered the maintenance phase. Induction cycles had 28 days and maintenance cycles had 84 days. Maintenance treatment was stopped once a (clinical) complete response (CR) and PB uMRD in 2 consecutive measurements were achieved, at progression, intolerable toxicity, or after the maximum number of 8 cycles of maintenance treatment. In the context of treatment discontinuation, clinical CRs included CRs that were not confirmed by imaging studies and/or a bone marrow biopsy. However, for the calculation of CR rates, only confirmed CRs (ie, by imaging and bone marrow examinations) were counted.
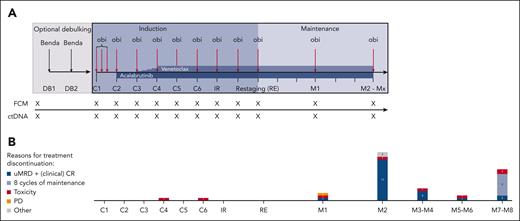
Treatment and sampling schedule. (A) Treatment discontinuations. (B) The number of patients discontinuing at certain time points is shown in the bar graphs. Patients who discontinued treatment according to the protocol due to achievement of undetectable MRD and a (clinical) CR are shown in dark blue; patients who completed a maximum of 8 cycles of maintenance are shown in blue. Patients who stopped treatment early because of toxicity are depicted in red and disease progressions are shown in yellow. Other reasons (gray) include “physician’s decision” (n = 1) and “allogeneic stem cell transplantation” (n = 1). Every x on the sampling schedule represents an FCM and ctDNA sample. Benda, bendamustine; C1, cycle 1; C2, cycle 2; C3, cycle 3; C4, cycle 4; C5, cycle 5; C6, cycle 6; DB, debulking cycle 1; DB2, debulking cycle 2; IR, initial response assessment; M1, maintenance staging 1; M2, maintenance staging 2; Mx, maintenance stagings 3 to 8; obi, obinutuzumab; PD, clinical disease progression; RE, final restaging.

Treatment and sampling schedule. (A) Treatment discontinuations. (B) The number of patients discontinuing at certain time points is shown in the bar graphs. Patients who discontinued treatment according to the protocol due to achievement of undetectable MRD and a (clinical) CR are shown in dark blue; patients who completed a maximum of 8 cycles of maintenance are shown in blue. Patients who stopped treatment early because of toxicity are depicted in red and disease progressions are shown in yellow. Other reasons (gray) include “physician’s decision” (n = 1) and “allogeneic stem cell transplantation” (n = 1). Every x on the sampling schedule represents an FCM and ctDNA sample. Benda, bendamustine; C1, cycle 1; C2, cycle 2; C3, cycle 3; C4, cycle 4; C5, cycle 5; C6, cycle 6; DB, debulking cycle 1; DB2, debulking cycle 2; IR, initial response assessment; M1, maintenance staging 1; M2, maintenance staging 2; Mx, maintenance stagings 3 to 8; obi, obinutuzumab; PD, clinical disease progression; RE, final restaging.
This study was conducted in accordance with the Declaration of Helsinki and the International Conference on Harmonization Good Clinical Practice Guidelines. The CLL2-BAAG trial was registered at ClinicalTrials.gov (identifier: NCT03787264). Written informed consent was provided before enrollment and the trial protocol was approved by the responsible health authorities and institutional review boards.
MRD samples were taken monthly during the induction phase (cycle 1-6) and 3-monthly in the MRD-guided maintenance phase. MRD was measured by conventional 4-color FCM from PB and ctDNA was assessed by digital droplet polymerase chain reaction (ddPCR) of patient/disease–specific variable-diversity-joining (VDJ) rearrangements and CLL–related somatic mutations in blood plasma.
ctDNA-based MRD assessment
Patient-specific VDJ assay
All ctDNA analyses were performed at the central laboratory of Cologne, Germany. For each sampling time point (Figure 1A), 20 mL of blood was collected in 2 PAXgene ccfDNA tubes (PreAnalytix, Hombrechtikon, Switzerland) and processed within 3 days of sample collection. Plasma and DNA extraction were performed according to previously published protocols.29 The patient-specific VDJ sequence was acquired by Sanger sequencing from the central laboratory in Ulm, Germany, and analyzed using the international immunogenetics information system (igmt.org). With the IDT PrimerQuest tool (idtdna.com) TaqMan assays were designed, intentionally spanning VDJ segment boundaries to gain specificity. ddPCR was performed on the QX200 ddPCR system (Bio-Rad, Hercules, CA) using previously published protocols. The reference gene RPP30 was used to calculate the proportion of CLL-specific ctDNA and to determine the lower limit of detection in each run (supplemental Table 13, available on the Blood website). Data analysis was performed with QuantaSoft (version 1.7.4.0917). Samples were counted as positive if the fluorescence signal in the FAM channel was similar to the FAM signal in the positive control and no signal in the negative control was observed.
CLL-related mutations
Plasma samples were also screened for CLL-related mutations using the same ddPCR and data analysis set up at baseline, cycle 1, cycle 4, initial response assessment (ie, end of cycle 6), final restaging (ie, end of cycle 8), and all maintenance and follow-up visits. In accordance with digital MIQE guidelines, the assay for a certain gene mutation was considered positive if 3 signals similar to the positive control were observed.30
FCM
At all MRD sampling time points (Figure 1A), 10 mL of blood was collected in EDTA tubes and analyzed in the central laboratory in Kiel, Germany, if received within 48 hours after collection. MRD was measured using a modified international standardized four-color FCM protocol.31 According to the international workshop on CLL (iwCLL) guidelines, uMRD was defined as <1 CLL cell per 10 000 leukocytes (<10–4), consequently potentially detectable CLL cell populations at levels <10–4 were labeled uMRD.32
Statistical methods
Baseline characteristics were reported using counts and percentages for categorical variables and descriptive statistics for continuous variables. The MRD rates at different time points were calculated with respect to the whole analysis population. MRD recurrence after the end of treatment was defined as the detection of FCM MRD ≥10–4 in PB and/or ctDNA in blood plasma after the achievement of uMRD by FCM and undetectable ctDNA. The Kaplan-Meier method was used for time-to-event analyses regarding PFS (defined as the time between registration and first disease progression or death), overall survival (OS; defined as the time between registration and death), time to next treatment (defined as the time between registration and start of first antileukemic treatment), and time to achieve uMRD (defined as the time between registration and first achievement of uMRD). All statistical analyses were conducted using SPSS v27 and R v4.1.1 software.
The study was approved by the institutional review boards of all participating sites.
Results
In total, 46 patients with relapsed/refractory CLL were included in this trial. Because of a violation of the study’s inclusion/exclusion criteria, 1 patient was excluded from the full analysis set (see the narrative in the supplement). Baseline characteristics are shown in supplemental Table 1. The median number of previous treatments was 1 (range, 1-4), and 18 patients (40%) had already received BTKi and/or venetoclax (BTKi, 8; venetoclax, 7; both, 3) before inclusion in this study. Of 11 patients with prior BTKi exposure, 2 had received continuous BTKi (ibrutinib and ibrutinib plus rituximab) before study inclusion; both stopped BTKi treatment due to intolerance and did not have BTK or PLCG2 mutations at study enrollment on central NGS-based testing, which was mandatory for BTKi-pretreated patients. Of the 11 BTKi-pretreated patients, 9 had received time-limited combinations before study inclusion, with treatment durations ranging from 9.1 to 34.0 months (supplemental Table 2). Of the 44 patients with available information, 14 (31.8%) had 17p deletions and/or TP53 mutations, and 34 (75.6%) had unmutated immunoglobulin heavy-chain variable region genes (IGHVs).
Treatment exposure
With a median observation time of 36.3 months (range, 12.0-45.8), all 45 patients have now finished study treatment in the CLL2-BAAG trial. Eighteen patients (40%) received a debulking with bendamustine before induction treatment. The median duration of treatment was 14.7 months (range, 6.1-32.9), and 2 patients (4.4%) discontinued treatment during cycles 4 and 6 of induction, both due to cytopenia. Following the MRD- and response-guided treatment approach, 25 of 43 patients with maintenance treatment (58.1%) discontinued therapy according to the protocol because of confirmed uMRD and achievement of a (clinical) CR. Nine patients (20.9%) completed the maximum number of 8 maintenance cycles due to persistent MRD and/or nonachievement of a (clinical) CR (Figure 1B). The median number of administered maintenance cycles was 2 (range, 1-8). In total, 9 patients (20.9%) discontinued maintenance treatment earlier than foreseen by the study protocol, 1 due to a Richter transformation, 6 due to adverse events (AEs) (drug eruption, COVID-19, pure red cell aplasia, neutropenia, scabies, and peripheral edema), 1 due to an allogeneic stem cell transplantation, and 1 due to the treating physician’s decision. Among the 8 patients who discontinued induction or maintenance treatment early due to AEs, 7 had uMRD and all had a partial response at the time point of discontinuation.
Efficacy
As reported in the primary efficacy analysis of this trial, the uMRD rate in PB at final restaging (ie, after ∼6 months of triple combination treatment) was 75.6% with 10 patients showing detectable MRD ≥10–4. In the course of maintenance treatment, 7 of these 10 patients (70%) achieved uMRD, 1 patient (10%) had a transformation to Hodgkin lymphoma, and 2 patients (20%) still had detectable MRD after 8 cycles of maintenance treatment (Figure 2A). Notably, both patients with persistent MRD after 8 cycles of maintenance remained progression-free. With a median observation time of 36.3 months, 42 of 45 patients (93.3%) achieved a uMRD <10–4 in PB at any time point during the study (best uMRD rate), including 17 of 18 patients (94.4%) previously exposed to venetoclax and/or a BTKi and 13 of 14 patients (92.9%) with TP53 aberrations (Figure 2B). The median time to achieve uMRD in PB was 5.4 months from registration, with 32 of the 45 patients (71.1%) already achieving uMRD during induction treatment (Figure 2C). The rate of CRs also improved with longer follow-up; although only 6 of 45 patients (13.3%) had a CR at final restaging, this number increased to 19 of 43 patients with at least 1 dose of maintenance treatment (44.2%) at the end of maintenance treatment. uMRD rates at final restaging and the best uMRD rates were similar in patients who received a debulking with bendamustine (72.2% and 94.4%) and patients who did not (77.8% and 92.6%, supplemental Tables 3 and 4).
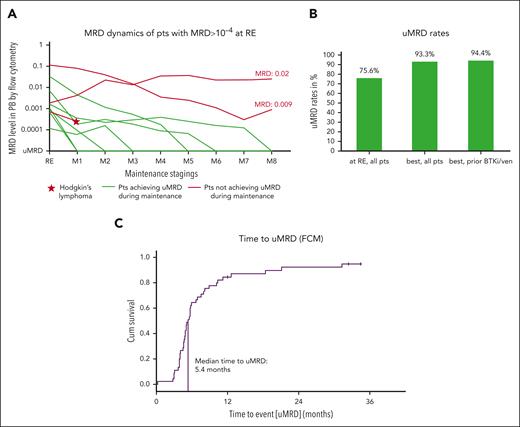
MRD dynamics and rates. (A) MRD levels of patients who still had detectable MRD in PB by FCM at RE. In 7 of 10 patients (green lines), undetectable MRD is achieved with ongoing maintenance; in 1 patient (red line and star), a transformation to Hodgkin lymphoma occurred while on treatment, and in 2 patients (red lines), MRD was still detected after 8 cycles of maintenance. (B) Undetectable MRD rates are shown for all patients at the RE. In addition, the best uMRD rates for all patients and those with prior exposure to BTK inhibitors and/or venetoclax are shown. (C) Time to achievement of undetectable MRD by FCM in PB. The median time to achievement of the uMRD is 5.4 months. M1-8, maintenance staging 1-8; Pts, patients; ven, venetoclax.

MRD dynamics and rates. (A) MRD levels of patients who still had detectable MRD in PB by FCM at RE. In 7 of 10 patients (green lines), undetectable MRD is achieved with ongoing maintenance; in 1 patient (red line and star), a transformation to Hodgkin lymphoma occurred while on treatment, and in 2 patients (red lines), MRD was still detected after 8 cycles of maintenance. (B) Undetectable MRD rates are shown for all patients at the RE. In addition, the best uMRD rates for all patients and those with prior exposure to BTK inhibitors and/or venetoclax are shown. (C) Time to achievement of undetectable MRD by FCM in PB. The median time to achievement of the uMRD is 5.4 months. M1-8, maintenance staging 1-8; Pts, patients; ven, venetoclax.
The median PFS was not reached, the estimated 3-year PFS rate for the whole analysis population was 85.0% (Figure 3). A similar PFS was shown for patients with TP53 aberrations (3-year PFS rate 85.7%; supplemental Figure 1) and patients previously exposed to BTKi and/or venetoclax (3-year PFS rate 94.4%; supplemental Figure 2). The estimated OS rate at 3 years was 93.8% (Figure 3), with 3 observed deaths in the study. The cause of death was COVID-19 in all 3 cases; all were assessed as unrelated to study treatment by the investigators and the time between the end of study treatment and death was 18.4, 24.2, and 25.2 months, respectively.
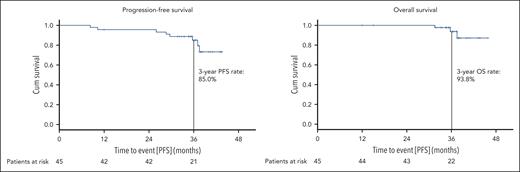
Progression-free and overall survival. PFS (A) and OS (B) are shown for the whole study population; patients at risk at the respective time points are listed below.
The median time to next treatment was not reached, with an estimated rate of 85.9% without subsequent treatment at 3 years. Subsequent treatments for patients with clinical progression were venetoclax/acalabrutinib 1 month after ibrutinib (n = 1), venetoclax/rituximab (n = 1), bendamustine/rituximab (n = 1), and adriamycin/bleomycin/vinblastine/dacarbacin (n = 1) in a patient with a transformation to Hodgkin lymphoma who later received acalabrutinib for his CLL. In addition, 2 patients continued acalabrutinib off study without fulfilling the iwCLL criteria for disease progression owing to high-risk genetics and/or insufficient MRD response. Furthermore, 1 patient with del(17p) and TP53 mutation who had already been treated with ibrutinib and venetoclax before study inclusion underwent allogeneic stem cell transplantation after maintenance cycle 2 and achievement of uMRD in PB.
Safety
At the time of the data cut (5 January 2024), 540 AEs were reported. Of these, 77 (14.3%) were CTC (Common Terminology Criteria for Adverse Events) grade 3, 24 (4.4%) were CTC grade 4, and 3 AEs were fatal (all COVID-19). Most AEs (n = 334, 61.9%) occurred during the induction phase, whereas on maintenance treatment, 88 AEs (16.3%) were reported (supplemental Table 9). In 86 cases (15.9%), AEs prompted an adjustment of study drugs; however, only 11 AEs (2.0%) led to permanent discontinuation of the study drug (supplemental Table 10). The most common AEs were infusion-related reactions in 24 patients (53.3%), thrombocytopenia in 20 patients (44.4%), diarrhea in 17 patients (37.8%), fatigue in 17 patients (37.8%), neutropenia in 15 patients (33.3%), COVID-19 in 13 patients (28.9%), nausea in 12 patients (26.7%), headache in 12 patients (26.7%), and rash in 11 patients (24.4%). One patient had an episode of atrial fibrillation (CTC grade 3) and no ventricular arrhythmias were reported.
ctDNA analyses
In total, 621 conventional MRD assessments by FCM and 619 plasma-based analyses by ddPCR were performed, resulting in 585 paired samples that were analyzed using both methods at the same time point. Missing samples were mostly because of insufficient quality of the shipped material or missed sampling at the sites. In 2 patients, no specific VDJ ddPCR assay could be designed because of unsuitable VDJ sequences. One of the 2 patients had a TP53 mutation that could be reliably tracked. Of the 585 paired FCM and ctDNA samples, 481 (82.2%) showed concordant results.
In total, 18 MRD recurrences were observed in the CLL2-BAAG trial. Of these, 5 were accompanied/followed by clinical progression and 13 were solely MRD recurrences not meeting the iwCLL criteria for clinical disease progression. In addition, 1 clinical progression (transformation to Hodgkin lymphoma) without MRD recurrence was observed. Among the 5 clinical progressions with MRD recurrence, 1 was Richter transformation (diffuse large B-cell lymphoma [DLBCL]). In this patient with DLBCL, patient-specific VDJ ctDNA was detected at the time point of DLBCL diagnosis, whereas conventional FCM indicated uMRD. In 2 of the 4 CLL-type progressions, ctDNA could be detected 7 and 17 months before clinical progression, which was also 6 and 14 months before FCM MRD became positive (patients P002 and P006; Figure 4). In patients P014 and P015, both FCM MRD and ctDNA recurred at the same time point. Three of the 4 clinical progressions were nodal progressions (patients P002, P006, and P014) with lymphocyte counts below 5 × 109/L (Figure 4).
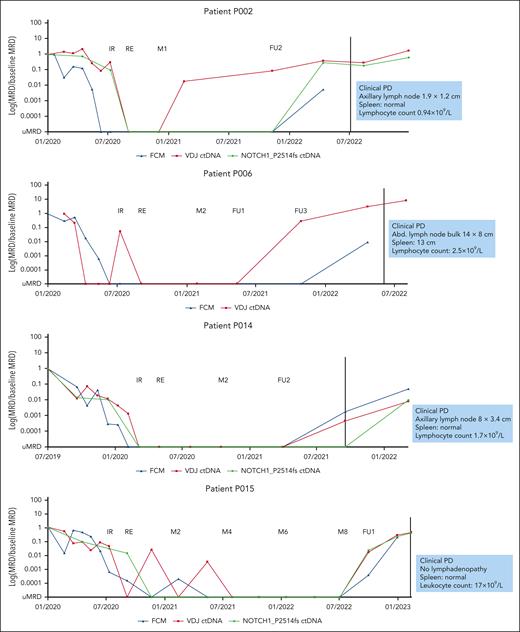
Detailed levels of FCM MRD and ctDNA for all 4 patients with CLL–type clinical disease progression. FCM MRD is shown in blue, ctDNA of patient-specific VDJ rearrangements in red, and in patients with available CLL-related mutations at baseline (P002, P0014, and P015), the ctDNA level of the somatic mutation is shown in green. In P002 and P006, ctDNA is detected several months before FCM MRD becomes positive and before clinical progression. In P014 and P015, ctDNA and FCM MRD both recur at the same time point. The characteristics of clinical progression are shown in blue boxes. FU1, follow-up staging 1; FU2, follow-up staging 2; FU3, follow-up staging 3; M2, maintenance staging 2; PD, clinical disease progression.

Detailed levels of FCM MRD and ctDNA for all 4 patients with CLL–type clinical disease progression. FCM MRD is shown in blue, ctDNA of patient-specific VDJ rearrangements in red, and in patients with available CLL-related mutations at baseline (P002, P0014, and P015), the ctDNA level of the somatic mutation is shown in green. In P002 and P006, ctDNA is detected several months before FCM MRD becomes positive and before clinical progression. In P014 and P015, ctDNA and FCM MRD both recur at the same time point. The characteristics of clinical progression are shown in blue boxes. FU1, follow-up staging 1; FU2, follow-up staging 2; FU3, follow-up staging 3; M2, maintenance staging 2; PD, clinical disease progression.
Among the 13 MRD recurrences without clinical progression, 1 showed FCM MRD and ctDNA recurrence at the same time point; in 3 cases, FCM MRD was already positive when ctDNA was still negative (range, 0-8 months difference) and in 9 patients with a solely molecular relapse, ctDNA was already detected when FCM still indicated uMRD (range, 0-8 months difference).
To better define constellations in which ctDNA was able to earlier detect (molecular) relapses, patients with clinical progression and/or MRD recurrence without clinical progression were grouped according to the method that first showed recurrence after the end of treatment (earlier detection by ctDNA/concordant/earlier detection by FCM) and relevant disease and treatment characteristics were added (Figure 5). Among the 12 patients with earlier detection by ctDNA, 10 patients (83.3%) had unmutated IGHV, 5 patients (41.7%) had del(11q), 6 patients (50.0%) were previously exposed to venetoclax/BTKi, and 5 of 9 evaluable patients (55.6%) had a complex karyotype. (Molecular) relapses detected by ctDNA occurred earlier (mean 7.2 months after the end of treatment) than in the group of patients with earlier detection by FCM (mean 20.3 months after the end of treatment). Of the 3 patients in whom relapse was first detected by FCM, 1 had unmutated IGHV (33.3%), whereas all had noncomplex karyotypes and no del(11q). In addition, no patient was previously exposed to venetoclax/BTKi and no clinical disease progressions were so far observed in this group.

MRD/ctDNA recurrences after end of treatment. All patients with MRD recurrence after or at the end of treatment by either FCM and/or ctDNA are shown, including patients with MRD recurrence and subsequent clinical disease progression. Each horizontal bar represents 1 patient (blue: prior exposure to BTK inhibitors and/or venetoclax; green: no prior BTK inhibitor/venetoclax) and starts at the end of study treatment (ie, time point 0 on the x-axis); that is, all MRD recurrences listed here occurred after study treatment was discontinued. Patients are grouped according to the method that first detected the MRD recurrence. The length of the bars shows the observation time for each patient between the end of treatment and the last available staging in months. The time points of ctDNA recurrence, FCM MRD recurrence, and clinical disease progression are shown for each patient if applicable. To clearly show the difference between FCM-based MRD recurrence and ctDNA recurrence, only the first positive results for each method are shown. Positive ctDNA or FCM results (ie, recurrences) that are followed by multiple negative results using the same method are not counted as MRD/ctDNA recurrences to reduce the risk of false-positive detection of molecular relapses. In the supplement, tables detailing all MRD/ctDNA results for each individual patient are shown. For each patient, certain clinical and genetic characteristics are shown on the left of the y-axis, next to the corresponding horizontal bar. Bulky disease was defined as at least 1 lymph node ≥5 cm in diameter on baseline computed tomography imaging. Benda, bendamustine; CKT, complex karyotype; Del(11q), deletion 11q; Del(13q), deletion 13q; Del(17p), deletion 17p; hCKT, highly complex karyotype; ven, venetoclax.

MRD/ctDNA recurrences after end of treatment. All patients with MRD recurrence after or at the end of treatment by either FCM and/or ctDNA are shown, including patients with MRD recurrence and subsequent clinical disease progression. Each horizontal bar represents 1 patient (blue: prior exposure to BTK inhibitors and/or venetoclax; green: no prior BTK inhibitor/venetoclax) and starts at the end of study treatment (ie, time point 0 on the x-axis); that is, all MRD recurrences listed here occurred after study treatment was discontinued. Patients are grouped according to the method that first detected the MRD recurrence. The length of the bars shows the observation time for each patient between the end of treatment and the last available staging in months. The time points of ctDNA recurrence, FCM MRD recurrence, and clinical disease progression are shown for each patient if applicable. To clearly show the difference between FCM-based MRD recurrence and ctDNA recurrence, only the first positive results for each method are shown. Positive ctDNA or FCM results (ie, recurrences) that are followed by multiple negative results using the same method are not counted as MRD/ctDNA recurrences to reduce the risk of false-positive detection of molecular relapses. In the supplement, tables detailing all MRD/ctDNA results for each individual patient are shown. For each patient, certain clinical and genetic characteristics are shown on the left of the y-axis, next to the corresponding horizontal bar. Bulky disease was defined as at least 1 lymph node ≥5 cm in diameter on baseline computed tomography imaging. Benda, bendamustine; CKT, complex karyotype; Del(11q), deletion 11q; Del(13q), deletion 13q; Del(17p), deletion 17p; hCKT, highly complex karyotype; ven, venetoclax.
Except for 1 PLCG2_S707F mutation that was detected in a patient ∼6 months after the end of maintenance treatment, no known resistance mutations (particularly in BTK or BCL2) were detected by plasma-based ddPCR analyses.
Discussion
With additional follow-up, the time-limited combination of acalabrutinib, venetoclax, and obinutuzumab after optional debulking with bendamustine demonstrated estimated PFS and OS rates of 85.0% and 93.8%, respectively, at 3 years in patients with relapsed/refractory CLL. Prolonged maintenance treatment in patients with persistent MRD led to deepening responses with the achievement of uMRD in most patients, with a best uMRD rate of 93.3% in PB. Extensive ctDNA-based analyses suggest an improvement in the early detection of (molecular) relapses when combining cell-based and cell-free approaches. No new safety signals were identified in the updated analysis of AEs and COVID-19 was responsible for all 3 fatal AEs in the study.
In general, but especially in patients with relapsed/refractory CLL, there are large differences in disease biology and dynamics, for example, due to the number of prior treatments and exposure to certain drug classes. In this diversely treated patient population, uniform treatment approaches, such as continuous monotherapies or fixed-duration combinations, might result in overtreatment with unnecessary toxicities and acquisition of resistance mutations or undertreatment with persistent disease at the end of treatment.33-39 The patient collective included in this trial represents a diversely treated relapsed/refractory CLL population with 1 to 4 prior CLL treatments, 40% of patients with prior BTKi/venetoclax exposure, 3 quarters had an unmutated IGHV, and almost one-third had TP53 aberrations. This variability was reflected by the different time points of uMRD achievement in this study. Although in most patients, uMRD was already achieved during induction treatment, a substantial number of patients who were still positive after 6 months of the triplet showed conversion to uMRD during maintenance treatment (n = 7). The presented MRD-guided treatment approach takes the different treatment requirements into account by stopping therapy early for those with good responses and extending treatment for those with persistent disease. This approach resulted in a median treatment duration of 14.7 months with broad variability (interquartile range, 12.8-27.8 months) reflecting differences in individual response dynamics. Following the MRD- and response-guided approach, uMRD was achieved in almost all patients during the course of the study, including those with prior BTKi/venetoclax and/or TP53 aberrations. With a median treatment duration of 14.7 months that is considerably shorter than the approved fixed-duration relapse therapy with venetoclax-rituximab, the triplet achieves uMRD and PFS rates that compare favorably with the MURANO trial: uMRD was achieved in 62.4% after 6 months of venetoclax-rituximab (CLL2-BAAG: 75.6% after 6 months of the triplet) which translated into a 3-year PFS rate of 71.4% (CLL2-BAAG: 85.0%). Although the patient populations in the MURANO and CLL2-BAAG trials are comparable with regard to most baseline characteristics, the type of previous treatments are not, as in MURANO, only a very small minority had received a BTK inhibitor, and no patient was previously exposed to venetoclax.11,40,41 The PFS observed with the CLL2-BAAG regimen also compares favorably to continuous BTKi monotherapy in the relapsed/refractory setting, that is, zanubrutinib (24-month PFS 78.4%, ALPINE trial), acalabrutinib, and ibrutinib (both, median PFS 38.4 months, ELEVATE-RR), with the obvious caveat of comparing data from phase 2 and phase 3 trials.12,42 Fixed-duration triplets or doublets of BTKi, venetoclax, and obinutuzumab have mostly been evaluated in treatment-naïve patients with CLL, but the MRD-adapted treatment approach from this trial also appears to lead to similar PFS rates when compared with fixed-duration first-line combinations from recent phase 3 trials, such as obinutuzumab-venetoclax (CLL14: 2-year PFS rate 88.2%; CLL13: 3-year PFS rate 87.7%), obinutuzumab-venetoclax-ibrutinib (CLL13: 3-year PFS rate 90.5%), or ibrutinib-venetoclax (GLOW: 2-year PFS rate 84.4%).2,3,9,43
In addition to these encouraging efficacy results in the relapsed/refractory setting, the relatively short median treatment duration in the CLL2-BAAG trial might also avoid the acquisition of resistance mutations, a known problem with continued BTKi therapy.34,35,44-47 With the caveat of limited follow-up and using an experimental plasma-based approach, only 1 patient showed a mutation in PLCG2 whereas no BTK and BCL2 mutations were observed in this trial, with all patients off-treatment for a median of 21.9 months. Based on several reports of patients re-exposed to time-limited venetoclax treatment, we assume that patients with disease progression after CLL2-BAAG might successfully be retreated with a similar regimen.18,48-50
All fatal AEs in this study were due to severe acute respiratory syndrome coronavirus 2 infections and were assessed as unrelated to the study treatment by the investigators. Given the timing of these fatal cases of COVID-19 between 18.4 and 25.2 months after the last dose of study treatment, we do not assume a relation of either of the study drugs with the COVID-19–related deaths and would not generally caution against using this combination in a patient population that is regularly exposed to severe acute respiratory syndrome coronavirus 2. The value of optional debulking with bendamustine has already been discussed in a previous article, in the current analysis we did not observe an impact of debulking on the attainment of uMRD at final restaging or throughout the study (best uMRD).18 In light of the currently established chemotherapy-free debulking strategies with BTKi, the bendamustine-based debulking approach used in this trial will most likely not play a role in most future clinical trials and practice.
A first prospective study on ctDNA-based MRD assessment in CLL demonstrated the general feasibility of the approach and suggested improved MRD detection by combining FCM– and ctDNA-based MRD methods.29 Moreover, ctDNA appeared to reflect residual disease across multiple compartments more adequately, as it did not seem to be as susceptible to fluctuations in CLL cell circulation in PB. In this study, we focused on the use of ctDNA-based analyses for early detection of clinical progressions and MRD recurrences.
One of the main findings of previous ctDNA analyses in CLL was that plasma-based MRD seemed to have advantages over FCM in patients with residual lymph nodes.29,51 In the present analysis, 3 of 4 CLL-type progressions were nodal relapses with low lymphocyte counts, and in 2 of these cases, ctDNA was positive considerably earlier than FCM-based MRD. In addition, 1 Richter transformation (DLBCL) occurred, in which ctDNA became positive at the time point of diagnosis, whereas no MRD could be detected by FCM, suggesting the detection of clonally related DLBCL ctDNA rather than CLL. With the limitation of very small numbers, these cases seem to underpin the results of the previous analyses.
With regard to MRD recurrences in this study, ctDNA was detected earlier in 12 patients, whereas FCM had advantages in 3 (Figure 5). Although absolute numbers were too small for meaningful confirmatory statistics, there seems to be a pattern regarding the composition of the 2 groups: among patients with earlier detection by ctDNA, a higher fraction of patients had high-risk genetics and del(11q) as a correlate for nodal CLL manifestation, more patients were previously exposed to BTKi/venetoclax, and MRD recurrence occurred earlier (mean 7.2 months after end of treatment) than in the group with earlier detection by FCM. Based on these results, it may be hypothesized that ctDNA-based analyses perform better in patients with more dynamic, high-risk disease.
However, given that FCM-based MRD was positive in 3 patients in whom no ctDNA could be detected, the use of ctDNA as a standalone method for MRD detection cannot be recommended based on this study. In future clinical trials, cell-free methods should only be used complementarily to a cell-based method to improve overall MRD detection and enable comparisons between the 2 approaches. Despite the dense sampling schedule, the large number of paired FCM and ctDNA samples (n = 585) and extensive longitudinal data that allow for the assessment of relapse detection, this study is limited by the absolute number of patients and consequently the low number of PFS and OS events, impeding correlative analyses of ctDNA with PFS and/or OS. In addition, because of limited follow-up, this data set only included patients with progressions or MRD conversions that occurred relatively early after the end of treatment, that is, in patients with a rather aggressive disease course. The certainly most important limitation of this ctDNA analysis is the comparison of the method with FCM-based MRD detection, with an established cutoff of 10–4. While both FCM and our plasma-based approach are inexpensive methods with a short turnaround time, facilitating quick MRD-based decision-making in this trial, FCM is not the most sensitive MRD method currently used in CLL. In most recent clinical trials, more sensitive cell-based methods (NGS- or allele-specific oligonucleotide PCR–based) that reliably achieve a lower limit of detection of at least 10–5 are used.43,52-55 To gain relevance in future trials and potentially in clinical practice, ctDNA-based approaches will have to demonstrate added value when compared with these currently most sensitive MRD methods. Considering the biological properties of ctDNA, the sensitivity of our currently rather narrow approach (VDJ plus several CLL-related mutations) might be substantially improved by using a broader sequencing method.25,26 It also needs to be stated that the earlier detection of a (molecular) relapse does so far not imply any clinical consequences, neither in the context of this clinical trial nor in routine practice. In conclusion, our ctDNA-based approach needs further validation and should currently only be used in clinical trials, ideally in combination with a standardized cell-based MRD method.
Ongoing analyses of a similar phase 2 trial (ClinicalTrials.gov identifier: NCT04515238) including a more sensitive cell-based and a broader plasma-based approach will build on this work and help to further define the role of ctDNA in CLL. A longer follow-up of this generation of phase 2 trials will ultimately enable correlative analyses of ctDNA with survival parameters and show whether the high rate of deep remissions observed in this trial will translate into long-term PFS.
Acknowledgments
The authors thank all patients, physicians, and trial staff at the sites for their participation in the study. The authors also thank Berit Falkowski, Aline Zey, Laura Miesen, and Anne Domonell, who worked as project managers; Olga Korf, Viktoria Monar, Anne Wosnitza, and Irene Preißler-Stodden, who worked as data managers; and Tanja Annolleck and Sabine Frohs, who worked as safety managers in the CLL2-BAAG trial.
This work was supported by the ERA PerMed program and the CLL-CLUE project. The trial was sponsored by the German CLL Study Group with financial support and study drug provision from Acerta/AstraZeneca and F. Hoffmann-La Roche.
Authorship
Contribution: M.F., J.W., A.G., S.R., M.H., and P.C. were responsible for the conception and design of the study; M.F., A.G., S.R., J. Stumpf, P.L., O.A.-S., F.S., A.-M.F., K.F., and P.C. were responsible for trial management; M.F., P.L., O.A.-S., C.S., E.T., J. Schetelig, P.D., S.B., M.R., S.S., B.E., M.H., and P.C. were responsible for recruitment and treatment of patients; M.F., A.G., S.R., and P.C. had access to the raw data; M.F. and P.C. did a central review of all clinical data; J.W., F.K., F.F., C.S., E.T., K.-A.K., M.R., A.S., M.B., and S.S. conducted the laboratory analyses; S.R. and A.G. performed the statistical analysis; M.F. and P.C. wrote the first draft of the manuscript; and all authors interpreted the data and reviewed and approved the manuscript.
Conflict-of-interest disclosure: M.F. received honoraria from AbbVie; and has received research funding (institution) from AbbVie, AstraZeneca, BeiGene, Janssen, and Roche. P.L. received grants and other supporting funding from Janssen-Cilag and other funding from AbbVie, AstraZeneca, and F. Hoffmann-La Roche. O.A.-S. received honoraria from Janssen-Cilag, Roche, Gilead Sciences, AbbVie, AstraZeneca, Adaptive Biotechnologies, and BeiGene; reports a consulting or advisory role in Roche, Janssen-Cilag, Gilead Sciences, and AbbVie; received research funding from BeiGene, Roche, AbbVie, and Janssen/Pharmacyclics; received travel, accommodations, and expenses from Roche, AbbVie, Gilead Sciences, and Janssen-Cilag. F.S. reports honoraria from AstraZeneca; research support from AstraZeneca; and travel support from Lilly Pharmaceuticals. A.-M.F. received research funding and honoraria from AstraZeneca; and travel grants from AbbVie. C.S. received honoraria from AbbVie, AstraZeneca, and Janssen. E.T. reports travel support from AbbVie, Janssen, and BeiGene; and honoraria for presentations and advisory board participation from AbbVie, AstraZeneca, BeiGene, Janssen, and Roche. J. Schetelig has received honoraria from AstraZeneca, Janssen, Bristol Myers Squibb (BMS), AbbVie, and BeiGene for participation in advisory boards, and received lecture fees from AstraZeneca, Janssen, BMS, AbbVie, BeiGene, Novartis, and Eurocept. P.D. reports a consultancy with AbbVie, AstraZeneca, bluebird bio, Gilead, Janssen, Novartis, Riemser, and Roche; and is on the speakers’ bureau for AbbVie, AstraZeneca, Gilead, Novartis, Riemser, and Roche. S.B. received payment or honoraria for lectures, presentations, speaker’s bureaus, manuscript writing, or educational events from Roche, AbbVie, Novartis, Becton Dickinson, Janssen, AstraZeneca, and Sanofi; and received grants or contracts from Janssen Cilag Neuss. K.F. reports honoraria and personal fees from AbbVie, AstraZeneca, and Roche; research funding from AbbVie, and Roche; and travel support from Roche. K.-A.K. reports a consultancy, speaker bureau fees, and research support from Janssen, Roche, and AbbVie. M.R. has received payments/honoraria for lectures/presentations from Janssen, AstraZeneca, and Roche; consulting fees from AbbVie, AstraZeneca, Janssen, and Roche; was supported for attending meetings/travel by AstraZeneca; and received grants/contracts from AbbVie and F. Hoffmann-La Roche. M.B. received personal fees from Incyte (advisory board); grants and personal fees from Amgen (advisory board, speakers bureau, travel support); and personal fees from Becton Dickinson, Janssen, and Pfizer (speakers bureau), all outside the submitted work. S.S. received advisory board honoraria, research support, travel support, and speakers’ fees from AbbVie, Amgen, AstraZeneca, Celgene, Gilead, GlaxoSmithKline, F. Hoffmann-La Roche, Janssen, Novartis, and Sunesis. B.E. was part of the speakers’ bureaus and participated in advisory boards from AbbVie, Janssen, and Roche. M.H. received honoraria, consulting fees, and grants from AbbVie, Roche, Janssen, AstraZeneca, Gilead, and BeiGene. P.C. received research funding from Acerta, AstraZeneca, BeiGene, F. Hoffmann-La Roche, Gilead, Janssen-Cilag, and Novartis; honoraria from AbbVie, AstraZeneca, BeiGene, BMS, F. Hoffmann-La Roche, and Janssen-Cilag; served on advisory boards by Acerta, AstraZeneca, BeiGene, Janssen-Cilag, and Novartis; and received travel support from AbbVie, AstraZeneca, BeiGene, F. Hoffmann-La Roche, Gilead, Janssen-Cilag, and Novo Nordisk. The remaining authors declare no competing financial interests.
The current affiliation of J.W. is Qiagen, Hilden, Germany.
Correspondence: Moritz Fürstenau, Department I of Internal Medicine Faculty of Medicine and University Hospital Cologne, Gleueler Str 176-178, 50935 Cologne, Germany; email: moritz.fuerstenau@uk-koeln.de; and Paula Cramer, German CLL Study Group (DCLLSG), Gleueler Str 176-178, 50935 Cologne, Germany; email: paula.cramer@uk-koeln.de.
References
Author notes
Data were presented in part as oral presentations at the 20th International Workshop on CLL, Boston, MA, 8 October 2023 and at the annual meeting of the American Society of Hematology, San Diego, CA, 9 December 2023.
Data can be shared upon request from the corresponding authors, Moritz Fürstenau (moritz.fuerstenau@uk-koeln.de) and Paula Cramer (paula.cramer@uk-koeln.de).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal