Key Points
The type of genetic variant is a predictive biomarker for disease severity and survival in WAS.
Less severe variants entail a later onset of disease-related complications but patients remain prone to morbidity and premature mortality.
Visual Abstract
Wiskott-Aldrich syndrome (WAS) is a multifaceted monogenic disorder with a broad disease spectrum and variable disease severity and a variety of treatment options including allogeneic hematopoietic stem cell transplantation (HSCT) and gene therapy (GT). No reliable biomarker exists to predict disease course and outcome for individual patients. A total of 577 patients with a WAS variant from 26 countries and a median follow-up of 8.9 years (range, 0.3-71.1), totaling 6118 patient-years, were included in this international retrospective study. Overall survival (OS) of the cohort (censored at HSCT or GT) was 82% (95% confidence interval, 78-87) at age 15 years and 70% (61-80) at 30 years. The type of variant was predictive of outcome: patients with a missense variant in exons 1 or 2 or with the intronic hot spot variant c.559+5G>A (class I variants) had a 15-year OS of 93% (89-98) and a 30-year OS of 91% (86-97), compared with 71% (62-81) and 48% (34-68) in patients with any other variant (class II; P < .0001). The cumulative incidence rates of disease-related complications such as severe bleeding (P = .007), life-threatening infection (P < .0001), and autoimmunity (P = .004) occurred significantly later in patients with a class I variant. The cumulative incidence of malignancy (P = .6) was not different between classes I and II. It confirms the spectrum of disease severity and quantifies the risk for specific disease-related complications. The class of the variant is a biomarker to predict the outcome for patients with WAS.
Introduction
Wiskott-Aldrich syndrome (WAS) is an inborn error of immunity (IEI) with X-linked inheritance pattern characterized by immunodeficiency, thrombocytopenia, eczema, and a predisposition to autoimmunity and malignancy.1 Due to its characteristic clinical presentation with thrombocytopenia and eczema, it is relatively easy to distinguish clinically from other rare monogenic IEIs.2 This explains why WAS was recognized early as a distinct, inherited clinical entity3,4 and its genetic cause identified in the 1990s.5 It was one of the first IEIs to be cured by allogeneic hematopoietic stem cell transplantation (HSCT)6 and treated by hematopoietic stem and progenitor cell gene therapy (GT).7 Nevertheless, WAS remains a multifaceted disorder posing very complex challenges to physicians and families alike.2,8
Patients affected by WAS carry hemizygous variants in the WAS gene and consequently express either no WAS protein (WASP) or reduced amounts of a defective protein.9 The penetrance, severity, and time of onset of clinical disease manifestations can be extremely variable between affected individuals and even within families.1,10-12 Some patients may suffer from life-threatening opportunistic infections and bleeding in early infancy and are prone to premature death in childhood unless treated curatively, whereas others may have a normal life span with mostly asymptomatic thrombocytopenia as the only symptom.10,13 A disease score ranging from 1 to 5 according to the severity of symptoms has been developed2 but is not well suited for therapeutic decision-making because it only describes disease severity at a given time point, whereas progression from milder to a more severe score can occur.10,14
Different therapeutic approaches are available for WAS, ranging from symptomatic treatment, prophylactic application of antimicrobial agents or immunoglobulin replacement, splenectomy, thrombopoietic agents, to definitive therapeutic options such as HSCT or GT.2,8,15-22 Anti-infective drugs or immunoglobulin replacement may prevent many severe infections but will not eliminate the risk of bleeding, autoimmunity, or malignancy. Splenectomy, and to a lesser degree thrombopoietic agents, can elevate the platelet count and reduce the risk of bleeding. Splenectomized patients are at a significant and persistent risk of life-threatening bacterial infections, in particular sepsis.10,15,21 HSCT has become increasingly successful and can completely cure the disease but carries a small but significant risk of mortality as well as short and long-term morbidity.17,18,22 Lentiviral GT results in at least partial correction of WASP expression across all hematopoietic cell lineages and leads to reversion or improvement of disease manifestations. Long-term effectiveness and safety still require additional follow-up, but no events of insertional mutagenesis have been observed with lentiviral GT for WAS after follow-up of up to 13 years.16,20,23
Previous studies have established a certain degree of genotype/phenotype correlation in WAS but demonstrated that even patients with milder variants are at a significant risk of severe disease-associated events.9-12 For patients with a classic WAS phenotype in childhood with a suitable stem cell donor, HSCT is the best available option and clearly indicated.17,18,24 On the contrary, it can be quite difficult to choose the optimal treatment strategy for patients with milder WAS phenotypes, considering risk and benefit because disease severity may aggravate during a patient’s life, and even clinically mild symptoms may have a significant negative impact on quality of life.25
Here, we provide a comprehensive overview of the distribution of disease burden in patients with WAS, assess their natural disease outcome, define the risk for specific disease-related events, and possibly describe genetic biomarkers that could be helpful in defining the most appropriate treatment strategy based on the risk profile of each patient with WAS.
Materials and methods
Data accrual
Pseudonymized case report forms, asking for retrospective data retrieved from patient records, were sent out to members of the Inborn Errors Working Party of the European Society for Blood and Marrow Transplantation and the European Society for Immunodeficiencies, as well as to the mailing list of the International Union of Immunological Societies. Additionally, centers known to treat patients with WAS and former collaboration partners were contacted directly. The cutoff date for data collection was 31 December 2014.
Patients
All 581 submitted case report forms were evaluated, and 577 patients could be enrolled in our study. Patients without genetic analysis of the WAS gene (2 patients) or with a variant in a different gene (WIP, 2 patients) were excluded. Patients with X-linked neutropenia caused by WAS variants were not part of this study.
Definitions
Disease-related complications were defined as follows: severe bleeding was defined as intracranial, gastrointestinal, or other life-threatening bleeding, or any bleeding episode requiring red blood cell transfusion; and severe infections were defined as sepsis, meningitis, pneumonia requiring respiratory support, systemic viral (viremia), or invasive fungal infections. For classification of current disease severity, we used the WAS score as previously published.2
For the purpose of this study, we divided the variants into 2 classes:
Class I: missense variants in exons 1 and 2 as well as the intronic hot spot variant c.559+5G>A, all of which had previously been defined as hot spot variants, often found in mildly affected patients and which are expected to allow for some WASP expression.9-12,26
Class II: all other WAS variants.
WASP expression was not systematically recorded or experimentally assessed in this study. WAS variants were curated according to the American College of Medical Genetics and Genomics (ACMG) criteria using the Franklin by Genoox tool.27,28
Karnofsky/Lansky scores were translated to Eastern Cooperative Oncology Group (ECOG) scores as suggested by Oken et al: Karnofsky/Lansky 100% = ECOG 0; 80% to 90% = ECOG 1; and ≤70% = ECOG ≤2.29
Statistical analysis
Kaplan-Meier survival estimates were compared using the log rank test. The survival analyses were censored at the time point of first HSCT or GT when indicated (all survival analyses except Figure 2A). Cumulative incidences for different events factored in only the first event of the specified category and were adjusted for competing risks. The mean incidences per patient-year were calculated for different time intervals, taking the patients at risk into account, counting each event of the specified category. Because those estimated mean incidences do not follow a normal distribution, the confidence intervals (CIs) were calculated with bootstrap techniques (percentiles of 1000 samples). All incidence analyses were censored at the time point of the first procedure (HSCT, GT, or splenectomy). All statistical analyses were performed with R version 3.5.3.30
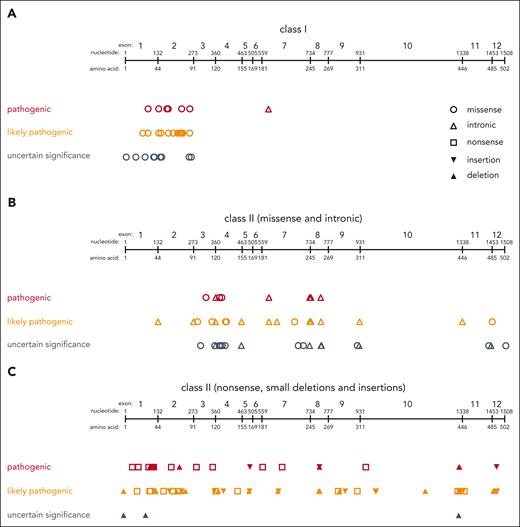
Classification of WAS variants. Distribution of class I variants (A) and class II variants (B-C) across the WAS gene. Variants are displayed as classified according to ACMG criteria as either “pathogenic” (red), “likely pathogenic” (orange), or “uncertain significance (VUS)” (gray). Variants classified as “likely benign” or “benign” are not shown.
Classification of WAS variants. Distribution of class I variants (A) and class II variants (B-C) across the WAS gene. Variants are displayed as classified according to ACMG criteria as either “pathogenic” (red), “likely pathogenic” (orange), or “uncertain significance (VUS)” (gray). Variants classified as “likely benign” or “benign” are not shown.
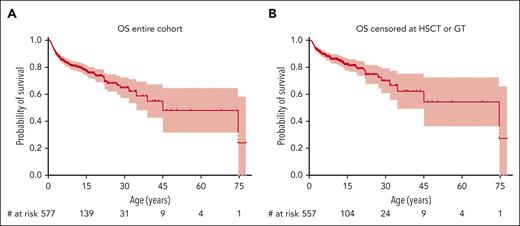
Overall Survival (OS). OS of the entire cohort (A) and of the entire cohort with patients censored at the time of HSCT or GT (B).
Overall Survival (OS). OS of the entire cohort (A) and of the entire cohort with patients censored at the time of HSCT or GT (B).
This retrospective chart-based survey was given a waiver by the ethics committee of the Ludwig Maximilian University of Munich, Munich, Germany. Individual centers gained local ethics committee approval for data transfer when it was required.
Results
Patients
We included 577 patients from 63 centers in 26 countries born between 1932 and 2014 in this study. The median age at diagnosis was 1.5 years (range, 0-68). Of these patients, 464 (80.4%) were alive at last follow-up, and the median age at last follow-up was 8.9 years (range, 0.3-71.1). This resulted in a total of 6118 reported patient-years.
The disease severity was variable across the cohort, with a WAS score at last follow-up or before the first procedure of 1 in 54 (9%), 2 in 144 (25%), 3 in 161 (28%), 4 in 109 (19%), or 5 in 86 patients (15%). HSCT had been performed in 255 patients (44%), splenectomy in 79 (14%), and GT in 14 (2%), and 42 (7%) had >1 of these procedures. More details of the cohort are reported in Table 1.
Characteristics of the cohort
| . | Entire cohort . | Class I variant . | Class II variant . | Variant not classified . | P∗ value . |
|---|---|---|---|---|---|
| Number of patients | 577 (100%) | 209 (100%) | 316 (100%) | 52 (100%) | |
| Region | |||||
| Europe | 278 (48.2%) | 106 (50.7%) | 139 (44.0%) | 33 (63.5%) | .1545 |
| United States | 113 (19.6%) | 67 (32.1%) | 42 (13.3%) | 4 (7.7%) | |
| Rest of world | 186 (32.2%) | 36 (17.2%) | 135 (42.7%) | 15 (28.8%) | |
| Russia | 54 (9.4%) | 12 (5.7%) | 41 (13.0%) | 1 (1.9%) | |
| China | 103 (17.9%) | 22 (10.5%) | 79 (25.0%) | 2 (3.8%) | |
| Brazil | 15 (2.6%) | 0 (0.0%) | 6 (1.9) | 9 (17.3%) | |
| Other | 14 (2.4%) | 2 (1.0%) | 9 (2.8) | 3 (5.8%) | |
| Age at diagnosis, y | |||||
| ≤2 | 309 (57.0%) | 76 (36.4%) | 199 (63.0%) | 34 (65.4%) | <.0001 |
| >2-5 | 118 (20.5%) | 46 (22.0%) | 59 (18.7%) | 13 (25.0%) | |
| >5-11 | 57 (9.9%) | 23 (11.0%) | 31 (9.8%) | 3 (5.8%) | |
| >11-18 | 35 (6.1%) | 24 (11.5%) | 10 (3.2%) | 1 (1.9%) | |
| >18 | 20 (3.5%) | 14 (6.7%) | 6 (1.9%) | 0 (0.0%) | |
| Not reported | 38 (6.6%) | 26 (12.4%) | 11 (3.5%) | 1 (1.9%) | |
| Median (range) | 1.5 (0.0-68.0) | 2.4 (0.0-68.0) | 1.3 (0.0-33.0) | 1.2 (0.0-11.3) | |
| Age at last follow-up, y | |||||
| ≤2 | 41 (7.1%) | 12 (5.7%) | 27 (8.5%) | 2 (3.8%) | .1343 |
| >2-5 | 89 (15.4%) | 27 (12.9%) | 55 (17.4%) | 7 (13.5%) | |
| >5-11 | 121 (21.0%) | 42 (20.1%) | 69 (21.8%) | 10 (19.2%) | |
| >11-18 | 86 (14.9%) | 40 (19.1%) | 37 (11.7%) | 9 (17.3%) | |
| >18 | 82 (14.2%) | 44 (21.1%) | 28 (8.9%) | 10 (19.2%) | |
| Not reported/dead (n, %) | 158 (27.4%) | 44 (21.1%) | 100 (31.6%) | 14 (26.9%) | |
| Median (range) | 8.9 (0.3-71.1) | 11.0 (0.3-68.0) | 6.7 (0.5-71.1) | 11.0 (1.5-42.0) | |
| Survival status at last follow-up | |||||
| Alive | 464 (80.4%) | 190 (90.9%) | 235 (74.4%) | 39 (75.0%) | <.0001 |
| Deceased | 113 (19.6%) | 19 (9.1%) | 81 (25.6%) | 13 (25.0%) | |
| WAS score at last follow-up or before first procedure | |||||
| 1 | 54 (9.4%) | 47 (22.5%) | 7 (2.2%) | 0 (0.0%) | <.0001 |
| 2 | 144 (25.0%) | 84 (40.2%) | 53 (16.8%) | 7 (13.5%) | |
| 3 | 161 (27.9%) | 40 (19.1%) | 98 (31.0%) | 23 (44.2%) | |
| 4 | 109 (18.9%) | 10 (4.8%) | 86 (27.2%) | 13 (25.0%) | |
| 5 | 86 (14.9%) | 19 (9.1%) | 60 (19.0%) | 7 (13.5%) | |
| Not reported | 23 (4.0%) | 9 (4.3%) | 12 (3.8%) | 2 (3.8%) | |
| Procedures | |||||
| Splenectomy | 79 (13.7%) | 31 (14.8%) | 40 (12.7%) | 8 (15.4%) | .5456 |
| HSCT | 255 (44.2%) | 44 (21.1%) | 177 (56%) | 34 (65.4%) | |
| GT | 14 (2.4%) | 4 (1.9%) | 10 (3.2%) | 0 (0%) | |
| >1 procedure | 30 (5.2%) | 4 (1.9%) | 21 (6.6%) | 5 (9.6%) | |
| No procedure | 263 (45.6%) | 134 (64.1%) | 114 (36.1%) | 15 (28.8%) | |
| Age at first procedure, y | |||||
| ≤2 | 115 (19.9%) | 13 (6.2%) | 88 (27.8%) | 14 (26.9%) | .0001 |
| >2-5 | 103 (17.9%) | 28 (13.4%) | 63 (19.9%) | 12 (23.1%) | |
| >5-11 | 40 (6.9%) | 15 (7.2%) | 17 (5.4%) | 8 (15.4%) | |
| >11-18 | 20 (3.5%) | 6 (2.9%) | 12 (3.8%) | 2 (3.8%) | |
| >18 | 9 (1.6%) | 4 (1.9%) | 5 (1.6%) | 0 (0.0%) | |
| No procedure or not reported, n (%) | 290 (50.3%) | 143 (68.4%) | 131 (41.5%) | 16 (30.8%) | |
| Median (range) | 2.3 (0.4-33.7) | 3.5 (0.5-33.7) | 2.1 (0.4-29.0) | 2.2 (0.4-13.8) |
| . | Entire cohort . | Class I variant . | Class II variant . | Variant not classified . | P∗ value . |
|---|---|---|---|---|---|
| Number of patients | 577 (100%) | 209 (100%) | 316 (100%) | 52 (100%) | |
| Region | |||||
| Europe | 278 (48.2%) | 106 (50.7%) | 139 (44.0%) | 33 (63.5%) | .1545 |
| United States | 113 (19.6%) | 67 (32.1%) | 42 (13.3%) | 4 (7.7%) | |
| Rest of world | 186 (32.2%) | 36 (17.2%) | 135 (42.7%) | 15 (28.8%) | |
| Russia | 54 (9.4%) | 12 (5.7%) | 41 (13.0%) | 1 (1.9%) | |
| China | 103 (17.9%) | 22 (10.5%) | 79 (25.0%) | 2 (3.8%) | |
| Brazil | 15 (2.6%) | 0 (0.0%) | 6 (1.9) | 9 (17.3%) | |
| Other | 14 (2.4%) | 2 (1.0%) | 9 (2.8) | 3 (5.8%) | |
| Age at diagnosis, y | |||||
| ≤2 | 309 (57.0%) | 76 (36.4%) | 199 (63.0%) | 34 (65.4%) | <.0001 |
| >2-5 | 118 (20.5%) | 46 (22.0%) | 59 (18.7%) | 13 (25.0%) | |
| >5-11 | 57 (9.9%) | 23 (11.0%) | 31 (9.8%) | 3 (5.8%) | |
| >11-18 | 35 (6.1%) | 24 (11.5%) | 10 (3.2%) | 1 (1.9%) | |
| >18 | 20 (3.5%) | 14 (6.7%) | 6 (1.9%) | 0 (0.0%) | |
| Not reported | 38 (6.6%) | 26 (12.4%) | 11 (3.5%) | 1 (1.9%) | |
| Median (range) | 1.5 (0.0-68.0) | 2.4 (0.0-68.0) | 1.3 (0.0-33.0) | 1.2 (0.0-11.3) | |
| Age at last follow-up, y | |||||
| ≤2 | 41 (7.1%) | 12 (5.7%) | 27 (8.5%) | 2 (3.8%) | .1343 |
| >2-5 | 89 (15.4%) | 27 (12.9%) | 55 (17.4%) | 7 (13.5%) | |
| >5-11 | 121 (21.0%) | 42 (20.1%) | 69 (21.8%) | 10 (19.2%) | |
| >11-18 | 86 (14.9%) | 40 (19.1%) | 37 (11.7%) | 9 (17.3%) | |
| >18 | 82 (14.2%) | 44 (21.1%) | 28 (8.9%) | 10 (19.2%) | |
| Not reported/dead (n, %) | 158 (27.4%) | 44 (21.1%) | 100 (31.6%) | 14 (26.9%) | |
| Median (range) | 8.9 (0.3-71.1) | 11.0 (0.3-68.0) | 6.7 (0.5-71.1) | 11.0 (1.5-42.0) | |
| Survival status at last follow-up | |||||
| Alive | 464 (80.4%) | 190 (90.9%) | 235 (74.4%) | 39 (75.0%) | <.0001 |
| Deceased | 113 (19.6%) | 19 (9.1%) | 81 (25.6%) | 13 (25.0%) | |
| WAS score at last follow-up or before first procedure | |||||
| 1 | 54 (9.4%) | 47 (22.5%) | 7 (2.2%) | 0 (0.0%) | <.0001 |
| 2 | 144 (25.0%) | 84 (40.2%) | 53 (16.8%) | 7 (13.5%) | |
| 3 | 161 (27.9%) | 40 (19.1%) | 98 (31.0%) | 23 (44.2%) | |
| 4 | 109 (18.9%) | 10 (4.8%) | 86 (27.2%) | 13 (25.0%) | |
| 5 | 86 (14.9%) | 19 (9.1%) | 60 (19.0%) | 7 (13.5%) | |
| Not reported | 23 (4.0%) | 9 (4.3%) | 12 (3.8%) | 2 (3.8%) | |
| Procedures | |||||
| Splenectomy | 79 (13.7%) | 31 (14.8%) | 40 (12.7%) | 8 (15.4%) | .5456 |
| HSCT | 255 (44.2%) | 44 (21.1%) | 177 (56%) | 34 (65.4%) | |
| GT | 14 (2.4%) | 4 (1.9%) | 10 (3.2%) | 0 (0%) | |
| >1 procedure | 30 (5.2%) | 4 (1.9%) | 21 (6.6%) | 5 (9.6%) | |
| No procedure | 263 (45.6%) | 134 (64.1%) | 114 (36.1%) | 15 (28.8%) | |
| Age at first procedure, y | |||||
| ≤2 | 115 (19.9%) | 13 (6.2%) | 88 (27.8%) | 14 (26.9%) | .0001 |
| >2-5 | 103 (17.9%) | 28 (13.4%) | 63 (19.9%) | 12 (23.1%) | |
| >5-11 | 40 (6.9%) | 15 (7.2%) | 17 (5.4%) | 8 (15.4%) | |
| >11-18 | 20 (3.5%) | 6 (2.9%) | 12 (3.8%) | 2 (3.8%) | |
| >18 | 9 (1.6%) | 4 (1.9%) | 5 (1.6%) | 0 (0.0%) | |
| No procedure or not reported, n (%) | 290 (50.3%) | 143 (68.4%) | 131 (41.5%) | 16 (30.8%) | |
| Median (range) | 2.3 (0.4-33.7) | 3.5 (0.5-33.7) | 2.1 (0.4-29.0) | 2.2 (0.4-13.8) |
χ2 test for the null hypothesis that the proportions of subjects in category x are equal in class I variant and class II variant subjects. x is the respective category in the same row. Subjects with missing values are not included in the analysis. All P values are unadjusted.
All patients in this cohort had a clinical diagnosis of WAS and a presumably disease-causing genetic variant in the WAS gene. In 52 patients, the WAS variant was confirmed yet not unambiguously reported. Of the 525 patients with clearly reported variants, 239 (46%) had a missense variant, 80 (15%) a nonsense variant, 90 (17%) an intronic variant, 76 (15%) a deletion, and 40 (8%) an insertion (Table 2). After curation according to ACMG criteria, 290 variants (55.2%) were classified as “pathogenic,” 170 (32.4%) as “likely pathogenic,” and 60 (11.4%) as “variants of uncertain significance (VUS)” (Figure 1; Table 2). Two (0.4%) were curated as “likely benign” and 3 (0.6%) as “benign.” Of these, 120 individual variants were, at the time of writing, not registered in ClinVar, 80 (66.7%) of which were found to have been previously published.9-12,31-47,48,49 Details of these variants and their curation according to the ACMG criteria are provided in supplemental Table 1, available on the Blood website.
Genetics
| Patients with exact genetic information, n (%) | 525 (100%) |
| Missense | 239 (45.5%) |
| Nonsense | 80 (15.2%) |
| Intronic | 90 (17.1%) |
| Deletion | 76 (14.5%) |
| Insertion | 40 (7.6%) |
| Classes of variants | 525 (100%) |
| Class I: missense in exon 1+2 or c.559+5G>A | 209 (39.8%) |
| Class II: other exact variants | 316 (60.2%) |
| Classification of variants according to ACMG criteria | 525 (100%) |
| Class I: missense in exon 1+2 or c.559+5G>A | 209 (39.8%) |
| Pathogenic | 160 (76.6%) |
| Likely pathogenic | 37 (17.7%) |
| VUS | 11 (5.3%) |
| Likely benign | 1 (0.5%) |
| Benign | — |
| Class II: other exact variants | 316 (60.2%) |
| Pathogenic | 130 (41.1%) |
| Likely pathogenic | 133 (42.1%) |
| VUS | 49 (15.5%) |
| Likely benign | 1 (0.3%) |
| Benign | 3 (0.9%) |
| Hot spots,∗n | |
| Class I | |
| c.C134T; p.Thr45Met | 15 |
| p.Val75Met/Gly/Leu | 60 |
| p.Arg86His/Leu/Gly/Cys | 59 |
| c.559 +5 G>A | 22 |
| Class II | |
| c.C121T ; p.Arg41X | 10 |
| c.C631T; p.Arg211X | 21 |
| c.777+1 G>A | 10 |
| Patients with exact genetic information, n (%) | 525 (100%) |
| Missense | 239 (45.5%) |
| Nonsense | 80 (15.2%) |
| Intronic | 90 (17.1%) |
| Deletion | 76 (14.5%) |
| Insertion | 40 (7.6%) |
| Classes of variants | 525 (100%) |
| Class I: missense in exon 1+2 or c.559+5G>A | 209 (39.8%) |
| Class II: other exact variants | 316 (60.2%) |
| Classification of variants according to ACMG criteria | 525 (100%) |
| Class I: missense in exon 1+2 or c.559+5G>A | 209 (39.8%) |
| Pathogenic | 160 (76.6%) |
| Likely pathogenic | 37 (17.7%) |
| VUS | 11 (5.3%) |
| Likely benign | 1 (0.5%) |
| Benign | — |
| Class II: other exact variants | 316 (60.2%) |
| Pathogenic | 130 (41.1%) |
| Likely pathogenic | 133 (42.1%) |
| VUS | 49 (15.5%) |
| Likely benign | 1 (0.3%) |
| Benign | 3 (0.9%) |
| Hot spots,∗n | |
| Class I | |
| c.C134T; p.Thr45Met | 15 |
| p.Val75Met/Gly/Leu | 60 |
| p.Arg86His/Leu/Gly/Cys | 59 |
| c.559 +5 G>A | 22 |
| Class II | |
| c.C121T ; p.Arg41X | 10 |
| c.C631T; p.Arg211X | 21 |
| c.777+1 G>A | 10 |
Defined as ≥10 patients with this variant.
Overall survival
Given the paucity of natural outcome data for patients with WAS in the present era, we performed a survival analysis of the entire cohort. The probability for overall survival (OS) was 78% (95% CI, 74-82) at age 15 years, 65% (95% CI, 58-73) at 30 years, and 55% (95% CI, 44-69) at 45 years (Figure 2A). Because potentially curative therapies such as HSCT or GT may significantly influence the natural course of disease either by curing the disease or by causing premature death, we performed an additional survival analysis in which patients treated by either HSCT or GT were censored on the date of that procedure (meaning their last follow-up was recorded on that date as “alive”). This resulted in an OS of 82% (95% CI, 78-87) at age 15 years, 70% (95% CI, 61-80) at 30 years, and 62% (95% CI, 50-77) at 45 years (Figure 2B). The approach of censoring at HSCT or GT was also used for all subsequent survival analyses.
Of the 113 patients who were reported deceased at the last follow-up, 31 (27%) had died from infectious causes, 26 (23%) from bleeding, 8 (7%) from malignancy, 4 (4%) from autoimmunity, 18 (16%) from HSCT-related events, and 26 (23%) from other or unknown causes (Table 3).
Causes of death
| All deceased patients | 113 (100%) |
| Bleeding | 26 (23.0%) |
| Intracranial | 16 |
| Pulmonary | 3 |
| Other/unknown | 7 |
| Infection | 31 (27.4%) |
| Bacterial | 13 |
| Sepsis | 8∗ |
| Pneumonia | 4 |
| Meningitis | 2 |
| Fungal | 2† |
| Viral | 8‡ |
| Other/unknown | 8 |
| Malignancy | 8 (7.0%) |
| Lymphoma | 6§ |
| Leukemia | 2‖ |
| Autoimmunity | 4 (3.5%) |
| AIHA | 3 |
| Colitis | 1 |
| Directly HSCT related | 18 (15.9%) |
| GVHD | 5 |
| VOD | 3 |
| Infection | 9 |
| Organ toxicity | 3 |
| Other/unknown | 2 |
| Other/unknown¶ | 26 (23.0%) |
| All deceased patients | 113 (100%) |
| Bleeding | 26 (23.0%) |
| Intracranial | 16 |
| Pulmonary | 3 |
| Other/unknown | 7 |
| Infection | 31 (27.4%) |
| Bacterial | 13 |
| Sepsis | 8∗ |
| Pneumonia | 4 |
| Meningitis | 2 |
| Fungal | 2† |
| Viral | 8‡ |
| Other/unknown | 8 |
| Malignancy | 8 (7.0%) |
| Lymphoma | 6§ |
| Leukemia | 2‖ |
| Autoimmunity | 4 (3.5%) |
| AIHA | 3 |
| Colitis | 1 |
| Directly HSCT related | 18 (15.9%) |
| GVHD | 5 |
| VOD | 3 |
| Infection | 9 |
| Organ toxicity | 3 |
| Other/unknown | 2 |
| Other/unknown¶ | 26 (23.0%) |
In the first category, the leading cause of death is given. Numbers in the subcategories may not add up because some patients had multiple reported causes of death.
AIHA, autoimmune hemolytic anemia; B-NHL, B Non-Hodgkin lymphoma; CMV, cytomegalovirus; DLBCL, diffuse large B-cell lymphoma; GVHD, graft-versus-host disease; T-ALL, T acute lymphoblastic leukemia; VOD, veno-occlusive disease.
Two pneumococcus, other organisms not reported.
One aspergillus, 1 organism not reported.
Three CMV, 5 organisms not reported.
One B-NHL, 1 DLBCL, others not specified.
Two T-ALL, both GT asociated, both relapsed after HSCT.
Contains deaths that occurred >6 months after HSCT and were not clearly attributable to HSCT.
Disease-related events
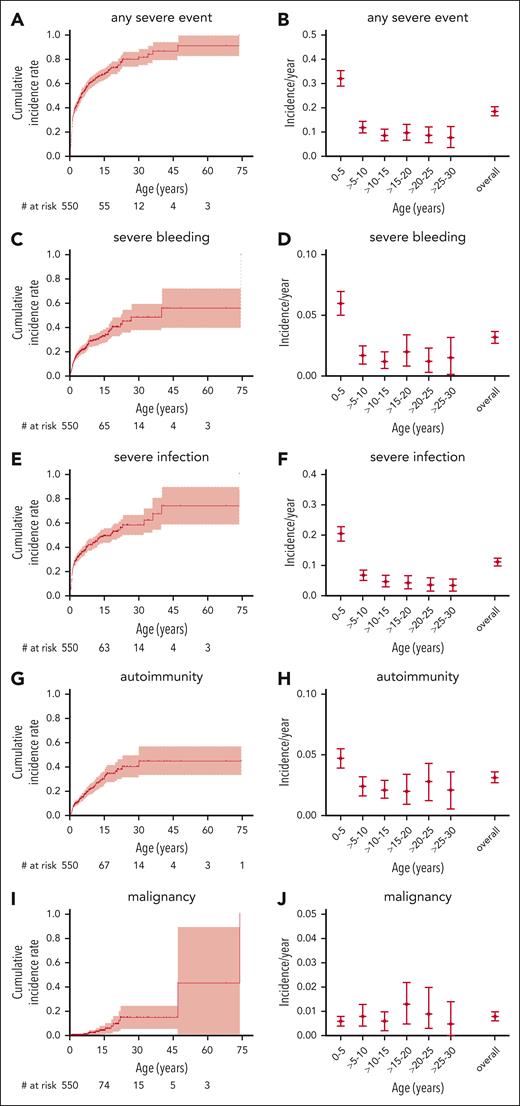
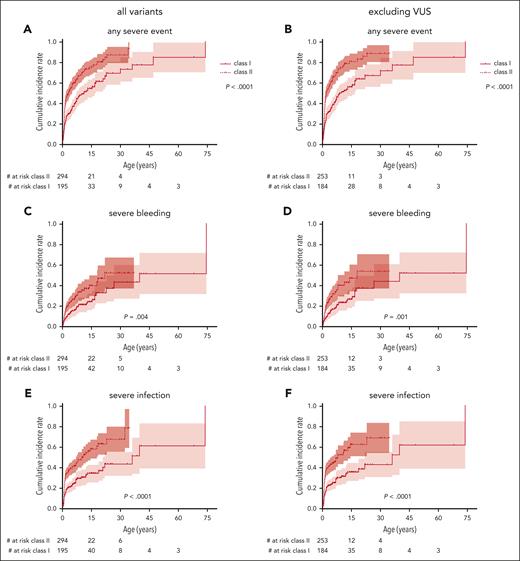
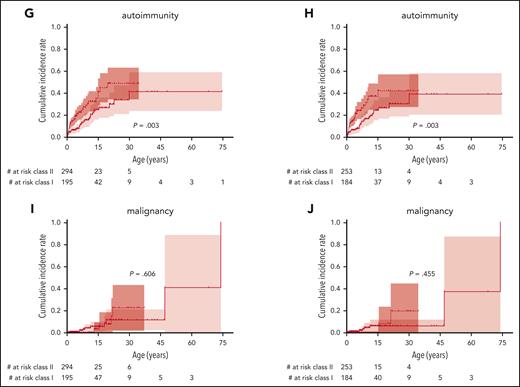
To better assess not only the mortality but also the morbidity caused by WAS, we analyzed the incidence of the most important disease-related events: severe bleeding, severe infection, autoimmunity, and malignancy, regardless of whether their outcome was fatal or not. The cumulative incidences of a first severe event at ages 15, 30, and 45 years were 33% (95% CI, 27-39), 49% (36-59), and 56% (36-70) for severe bleeding; 49% (43-55), 58% (49-66), and 74% (53-86) for severe infection; 30% (23-36), 40% (30-49), and 45% (31-56) for autoimmunity; and 4% (1-7), 15% (5-24), and 15% (5-24) for malignancy, respectively (Figure 3). For cumulative incidence analyses of events, the follow-up was also censored at the time of any first procedure (HSCT, GT, or splenectomy).
Severe disease-related events. Cumulative incidence and incidence per patient-year for severe disease-related events: any severe event (A-B), severe bleeding (C-D), severe infection (E-F), autoimmunity (G-H), and malignancy (I,J). Follow-up censored at the time of first procedure. Only the first event of a category was counted for cumulative incidence, whereas all events of a specific category were counted for incidence per year.
Severe disease-related events. Cumulative incidence and incidence per patient-year for severe disease-related events: any severe event (A-B), severe bleeding (C-D), severe infection (E-F), autoimmunity (G-H), and malignancy (I,J). Follow-up censored at the time of first procedure. Only the first event of a category was counted for cumulative incidence, whereas all events of a specific category were counted for incidence per year.
We calculated per-patient-year incidences of disease-related events to assess morbidity during a specific age period. The overall incidence of any severe event was 0.19 (0.17-0.20) per patient-year, including 0.03 per patient-year (0.03-0.04) for severe bleeding, 0.11 per patient-year (0.10-0.13) for severe infection, 0.03 per patient-year (0.03-0.04) for autoimmunity, and 0.01 per patient-year (0.01-0.01) for malignancy. Of note, bleeding, infection, and autoimmunity had a higher observed incidence in the first 5 years of life and then returned to a steady level for the next 25 years of life, whereas malignancy was equally frequent during any age period (Figure 3B,D,F,H,K). This may, at least partially, be explained by the fact that many severely affected patients had undergone HSCT in early childhood and were therefore censored from the analysis at later time points. Nevertheless, these data give an approximation of the true incidence of severe disease-related events during a specific age period in patients with WAS in the absence of a potentially definitive procedure such as HSCT or GT.
Class of the variant predicts outcome
Because the variability in disease severity in WAS can make it challenging to counsel families with respect to the most appropriate treatment choice, we intended to establish genotype as a predictive biomarker of disease outcome. For the purpose of this study, we grouped together all missense variants in exons 1 and 2 as well as the variant at the variant splice site c.559+5G>A, which generally allow for some (reduced) WASP expression, under the term “class I variants.” These types of variants had previously been defined as hot spot variants often found in mildly affected patients.9-12,26 All other WAS variants were grouped as “class II variants” (Table 2). Patients who had genetically confirmed WAS, but in whom the exact variants were not reported and those with “benign” or “likely benign” variants were excluded from this part of the analysis.
OS censored for HSCT and GT in patients with class I variants was significantly better than that of patients with class II variants: 93% (89-98) vs 71% (62-81) at 15 years, 91% (86-97) vs 48% (34-68) at 30 years and 84% (70-100) vs 41% (26-66) at 45 years, respectively (P < .0001; Figure 4A). When being more rigorous and also excluding patients with VUS, OS was also significantly better for patients with class I variants (P < .0001; Figure 4B).
OS by class of variant. (A) OS, comparing those with either a missense variant in exons 1+2 or the c.559+5G>A intronic variant (class I) with all others (class II). (B) Same analysis excluding patients with VUS according to ACMG criteria. Variants classified as “likely benign” or “benign” were excluded from this analysis.
OS by class of variant. (A) OS, comparing those with either a missense variant in exons 1+2 or the c.559+5G>A intronic variant (class I) with all others (class II). (B) Same analysis excluding patients with VUS according to ACMG criteria. Variants classified as “likely benign” or “benign” were excluded from this analysis.
The cumulative incidence of the first severe event was also significantly higher in patients with class II variants than those with class I variants, with 75% (67-81) vs 55% (45-63) at 15 years and 87% (76-93) vs 70% (56-79) at 30 years, respectively (P < .0001; Figure 5A). Accordingly, severe bleeding episodes, severe infections, and autoimmunity, but not malignancy, occurred significantly earlier in life in patients with class II variants (Figure 5C,E,G,I). Excluding patients with VUS, severe bleeding episodes, severe infections, and autoimmunity, but not malignancy, also occurred significantly earlier in patients with class II variants (Figure 5B,D,F,H,K). Patients with class II variants were diagnosed earlier in life, had a higher WAS score, and received their first procedure at a younger age (P ≤ .0001; Table 1). Yearly incidence rates for severe events are provided in supplemental Figure 2.
Incidence of severe events by class of variant. Cumulative incidence of the first severe disease-related event comparing those with class I variants (missense variant in exons 1+2 or the c.559+5G>A intronic variant) with class II variants (all other variants) including all patients (left) or excluding patients with VUS (right): any severe event (A-B), severe bleeding (C-D), severe infection (E-F), autoimmunity (G-H), and malignancy (I,J). Variants classified as “likely benign” or “benign” were excluded from this analysis.
Incidence of severe events by class of variant. Cumulative incidence of the first severe disease-related event comparing those with class I variants (missense variant in exons 1+2 or the c.559+5G>A intronic variant) with class II variants (all other variants) including all patients (left) or excluding patients with VUS (right): any severe event (A-B), severe bleeding (C-D), severe infection (E-F), autoimmunity (G-H), and malignancy (I,J). Variants classified as “likely benign” or “benign” were excluded from this analysis.
In summary, severe events occurred in both variant groups at comparable overall cumulative incidences but significantly later in life in those patients with class I variants.
Discussion
To our knowledge, this study describes the largest cohort of patients with WAS ever collected. In many previous studies, either patients with a distinct disease phenotype (“mild,” “classic,” or “early severe”)10,13 or just 1 therapeutic approach (usually HSCT or GT) were analyzed.16-18,22 In contrast, this cohort includes patients with all grades of disease severity and thus enabled us to give a comprehensive overview over the distribution of disease burden in patients with WAS and assess their natural disease outcome.
A certain degree of genotype/phenotype correlation in WAS has been known for a while.9,11,12 In this study, we identified a group of WAS genetic variants, which we termed class I, as predictive biomarkers for a less severe disease phenotype. Nevertheless, even these patients have a high probability of severe disease-related complications, which, on average, occur a few years later in life than that of patients with class II variants, but both patient groups have a significant risk of premature death. Our data advocate not to call class I variants “mild,” because that would imply a perpetuous benign course of disease, which is not the case for many patients. This seemingly stands in contrast to a cohort of patients with X-linked thrombocytopenia (XLT) with normal life expectancy that we previously reported.10 However, in the latter study, only patients with an already known mild disease course were included and retrospectively assessed, and even those had a significant incidence of disease-related events but much less frequent than the class I cohort described here.10 Approximately 73% of patients in that study had class I variants.10
Our findings are significant because they potentially allow for treating physicians to counsel families on the expected disease severity based on the type of variant that is usually known at the time of diagnosis or shortly after. Given the excellent OS after HSCT or GT,16-18,23 this knowledge opens the discussion toward offering curative treatments even to patients carrying a class I variant who do not, or maybe not yet, exhibit the full clinical spectrum of WAS disease manifestations. Currently, patients with a WAS score of 1 or 2 are labeled as XLT and often not considered for immediate curative therapy, which can be problematic if they develop life-threatening complications at an older age when permanent organ damage makes HSCT a risky procedure.50 On the contrary, the decision to perform transplantation in a young child with “mild” WAS has to be carefully weighed against the long-term risks of the procedure and discussed with the family, especially if a well-matched donor is not available. Our data suggest that the label “XLT” should be used with caution or even abandoned in favor of calling all patients with presumably disease-causing WAS variants “WAS patients” to encourage a genetics-based and more pragmatic therapeutic approach. Along the same lines, these data emphasize that the established “WAS score” should only be used to describe current disease severity but not as a basis for treatment decisions, because the score does not properly reflect the possibility of progression in disease severity. We also propose that the most suitable therapeutic approach should be reevaluated and set in context with the available scientific evidence for every patient with WAS at regular time intervals.
The categorization of WAS variants as class I or II was not arbitrary. We grouped missense variants located in exon 1 or 2, which have been reported to result in a reduced amount of WASP expression, into class I. One additional intronic variant that results in transcription of multiple splicing products, including normal WASP, previously identified as a hot spot for a less severe disease course, was also added to class I.9-12 Nevertheless, we cannot exclude that other WAS variants exist that are less detrimental than those labeled class II but have not been identified as such because they are rare.
All reported WAS variants in this cohort were classified according to the ACMG criteria. The variants of 5 patients were classified as “likely benign” or “benign.” Three of these variants were previously reported in other patients with WAS.10,34,35 All 5 had classical WAS features with thrombocytopenia, eczema, and WAS scores between 2 and 5, but they were, nevertheless, excluded from the analysis of genotype/phenotype correlation. Without access to original patient material, we are unable to decipher whether these variants are in fact pathogenic or whether these patients may have had additional deleterious variants in WAS that could not be detected or had a variant in another gene resulting in a phenocopy of WAS, also considering that the genetic analysis of some of these patients was performed in the early 2000s or before.
Although “benign” or “likely benign” variants have a sufficient probability to be not disease causing, this is not necessarily the case for VUS. They have to be carefully interpreted in conjunction with clinical evidence, and adequate variant curation is important. Therefore, we performed 2 separate analyses of genotype/phenotype correlation: one including patients with VUS and a more rigorous one excluding them. The results were very similar with regard to the difference between classes I and II with a slightly more severe phenotype for patients with known pathogenic and likely pathogenic variants. To make the variant curation that we performed more helpful for clinicians, those variants that are currently not described in ClinVar were provided with their respective curation criteria as a reference in supplemental Table 1. Many of these were reported for multiple patients in our cohort and/or had been previously reported in WAS.9-12,31-47
It is important to note that other factors besides the underlying variant can influence the individual disease course of a patient with WAS, such as somatic reversions, possibly disease-modifying genes, or administration of anti-infectives or thrombopoietic agents.19,21,51 Furthermore, the absolute numeric values for incidences of events in this study need to be interpreted cautiously, not only because of possible reporting bias, but also due to the fact that we censored the analysis after HSCT or GT, which are often carried out at a young age. The number of patients at risk tend to get very low at ages >15 years, and the incidences of disease complications at higher ages may therefore be underestimated because patients with more severe phenotypes received transplantation early and were excluded from the analysis after their HSCT or GT date. The same caveat applies to all Kaplan-Meier graphs in this article, in which the number of patients beyond 30 years of follow-up are small, thus resulting in wide CIs. It is important to note that this survey contains historic data, which preclude generalizing the findings to the current era with presumably improved supportive care and a lower threshold to proceed to HSCT in current times. There are other limitations to an international, retrospective chart-based analysis such as this, in which it is impossible to control for reporting bias or to monitor source data. The detail of HSCT-related data collected in this survey was very limited by design and not a focus of this study and therefore not reported here. There was no significant difference in OS between patients who underwent HSCT and those who did not (supplemental Figure 1A), but this comparison using the Kaplan-Meier method is hampered by significant bias, because severely affected patients are more likely to undergo HSCT early, and the transplant-related mortality will be higher in older patients. Survival after HSCT was 80% at 2 years and plateaued after that (supplemental Figure 1B). Two large, more recent cohorts from the United States and Europe that evaluated the outcomes of HSCT in WAS were published in 2020 and 2022, respectively, showing improved survival after HSCT regardless of donor source.17,18
In conclusion, this study demonstrates that the type of WAS variant is a valid biomarker, predictive of the expected disease severity of patients with WAS. Patients with the less severe class I variants can, on average, expect a later onset of severe WAS-related complications but remain prone to morbidity and premature mortality, justifying the evaluation for early definitive therapy in this group of patients with WAS. These findings shall help to improve the counseling of families with patients with WAS.
Acknowledgments
The authors thank Stephan Bischofberger (Staburo) for expert statistical advice.
This study was supported in part by the Wiskott-Aldrich Foundation, the Israeli Wiskott Aldrich Syndrome Association, and GlaxoSmithKline (M.H.A.). I.M. is a senior clinical investigator at Fonds Wetenschapelijk Onderzoek (FWO) Vlaanderen and is supported by KU Leuven C1 Grant (C16/18/007), FWO (grants G0C8517N, G0B5120N, and G0E8420N), and the Jeffrey Modell Foundation. M.H.A. and I.M. are representatives of reference centers of the European Reference Network on Rare Immunodeficency, auToinflammatory and Autoimmune diseases.
Authorship
Contribution: M.H.A., T.C.V., and J.S.G. designed the research and collected the data; M.H.A., T.C.V., J.S.G., and H.B. analyzed the data; all authors except H.B. provided original patient data; and all authors reviewed the final manuscript and consented to its submission.
Conflict-of-interest disclosure: I.M. reports research support from CSL Behring and being on a scientific advisory board for Boehringer-Ingelheim. M.H.A. reports research support by GlaxoSmithKline and Orchard and serving on a scientific advisory board for CSL Behring. The remaining authors declare no competing financial interests.
Correspondence: Michael H. Albert, Dr von Hauner University Children's Hospital, Lindwurmstr 4, 80337 Munich, Germany; email: malbert@med.lmu.de.
References
Author notes
Deidentified summary data are available on request from the corresponding author, Michael H. Albert (malbert@med.lmu.de).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal