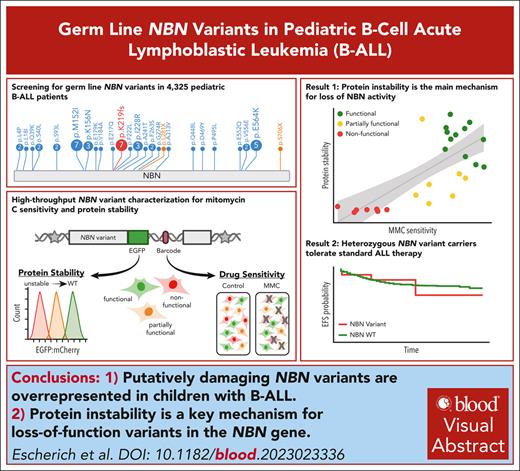
Heterozygous carriers of deleterious germ line NBN variants are at risk of B-ALL development.
Protein instability is the main mechanism for loss-of-function variants in the NBN gene.
Visual Abstract
Biallelic mutation in the DNA-damage repair gene NBN is the genetic cause of Nijmegen breakage syndrome, which is associated with predisposition to lymphoid malignancies. Heterozygous carriers of germ line NBN variants may also be at risk for leukemia development, although this is much less characterized. By sequencing 4325 pediatric patients with B-cell acute lymphoblastic leukemia (B-ALL), we systematically examined the frequency of germ line NBN variants and identified 25 unique, putatively damaging NBN coding variants in 50 patients. Compared with the frequency of NBN variants in gnomAD noncancer controls (189 unique, putatively damaging NBN coding variants in 472 of 118 479 individuals), we found significant overrepresentation in pediatric B-ALL (P = .004; odds ratio, 1.8). Most B-ALL–risk variants were missense and cluster within the NBN N-terminal domains. Using 2 functional assays, we verified 14 of 25 variants with severe loss-of-function phenotypes and thus classified these as nonfunctional or partially functional. Finally, we found that germ line NBN variant carriers, all of whom were identified as heterozygous genotypes, showed similar survival outcomes relative to those with wild type status. Taken together, our findings provide novel insights into the genetic predisposition to B-ALL, and the impact of NBN variants on protein function and suggest that heterozygous NBN variant carriers may safely receive B-ALL therapy. These trials were registered at www.clinicaltrials.gov as #NCT01225874, NCT00075725, NCT00103285, NCI-T93-0101D, and NCT00137111.
Introduction
B-cell acute lymphoblastic leukemia (B-ALL) is the most common cancer in children, and there is growing evidence for the inherited basis of ALL susceptibility.1 The majority of leukemia risk genes identified thus far are involved in either lymphoid differentiation (eg, ETV6, PAX5, IKZF1, and TCF3)1-5 or cell cycle and apoptosis signaling (eg, CDKN2A and TP53).6,7 DNA-damage repair has also been implicated in the pathogenesis of both lymphoid and myeloid leukemias. Germ line pathogenic variants in DNA repair genes interfere with the correction of DNA double-strand breaks (ataxia telangiectasia1 and Nijmegen breakage syndrome [NBS]8), single-strand breaks (constitutional mismatch repair deficiency),9 or interstrand crosslinks (Fanconi anemia).10 In these conditions, the failure of DNA repair leads to genome instability and thus an increased risk of hematological disorders.1
NBS is an autosomal-recessive condition caused by a biallelic loss-of-function mutation in the NBN gene.8 About 90% of patients with NBS are homozygous for the frameshift mutation, p.K219fs (c.657_661delACAAA), noted to be a founder mutation in populations of Eastern European (Slavic) descent.11,12 The NBN protein functions as a sensor for DNA double-strand breaks and an adapter for the downstream repair signaling.11 The N-terminus contains a Forkhead-associated and 2 breast cancer C-terminus domains (FHA-BRCT-repeat domain). This region allows interaction with the mediator of DNA-damage checkpoint 1 (MDC1) and phosphorylated histone H2AX, both of which accumulate at the site of DNA damage.13,14 The NBN C-terminus harbors the MRE11/RAD50 and ataxia telangiectasia mutated (ATM) protein kinase interaction sites, and thus implicated in cell cycle control, DNA repair, and apoptosis signaling.15,16 Patients with NBS develop characteristic phenotypes including immunodeficiency, radiosensitivity, and cancer susceptibility.8,17 The cumulative risk of developing cancer during childhood among patients with NBS is up to 70%, and lymphoid malignancies (B- or T-linage subtypes) are by far most frequently diagnosed.18,19
Heterozygous carriers of pathogenic NBN variants are clinically asymptomatic but are still believed to be at an increased risk for cancer development.20 Two large studies (with >34 00021 and >39 00022 patients with cancer) confirmed pan-cancer association with heterozygous NBN p.K219fs variant carriers, who are particularly prone to the development of breast cancer, prostate cancer, leukemia, and lymphoma.23,24 Besides truncating NBN variants, more than 160 missense germ line NBN variants have been identified,21 including the NBN variant p.I171V noted in pediatric B-ALL.25-27 Other germ line NBN variants (eg, p.A32fs, p.S93L, p.D95N, p.V210F,p.R215W, and p.K233fs) were described in patients with B-ALL, but with conflicting evidence regarding their impact on leukemia risk.23,25,26,28-30
Although there are a growing number of NBN variants detected in patients with cancer, efforts to assess their association with B-ALL predisposition are limited. In addition, the functional consequences of these variants remain largely uncharacterized, thus hampering clinical interpretation of variant pathogenicity. To address these challenges, we comprehensively screened for germ line NBN variants in 4325 pediatric patients with B-ALL, experimentally characterized these variants using 2 phenotyping assays, and evaluated their association with B-ALL characteristics and treatment outcomes.
Materials and methods
Patient cohort
It included 4325 patients enrolled in Children’s Oncology Group (COG) P9900 (NCT01225874), AALL0232 (NCT00075725), AALL0331 (NCT00103285) and St. Jude Total Therapy XIIIA, XIIIB (NCI-T93-0101D), and XV (NCT00137111) clinical trials for newly diagnosed patients with B-ALL were included for NBN-targeted sequencing.31-35 The study was approved by institutional review boards at St. Jude Children’s Research Hospital and COG member institutions, and informed consent was obtained from parents, guardians, or patients, as appropriate.
NBN-targeted sequencing
It was performed following procedures described previously.2,7,36 Briefly, genomic DNA was extracted from bone marrow or peripheral blood samples of pediatric patients with B-ALL obtained during remission. Illumina dual-indexed libraries were generated from patient germ line DNA and pooled in sets of 96 before hybridization with customized Roche NimbleGene SeqCap EZ probes (Roche NimbleGen, Inc, Madison, WI ) to capture NBN genomic region. Quantitative polymerase chain reaction was used to define the appropriate capture product titer necessary to efficiently populate an Illumina HiSeq 2000 flow cell for paired-end 2 × 100 bp sequencing. Whether or not a patient was included in the NBN genetic study was determined only by consent to participate in research, specimen availability, and sequencing quality, as described previously.7
NBN−/− HEK293T landing pad cellular model and NBN variant characterization
NBN gene knockout single clone was generated from HEK293T landing pad (LP) cells37-39 transiently transduced with Cas9 and single guide RNA targeting NBN (NBN−/− HEK293T LP cells). Details of single guide RNA and genotyping primers can be found in supplemental Table 5 (available on the Blood website). Next, NBN variants were integrated into the attachment Phage (attP) locus of NBN−/− HEK293T LP cells via Bxb1-mediated recombination.37,39 Cells with successful recombination were identified by flow cytometry as mCherry-positive and blue fluorescent protein (BFP)-negative (mCherry+BFP−) population. Subsequently, successfully recombined cells were used for NBN variant characterization.
NBN variant protein stability was tested after each variant was separately expressed in NBN−/− HEK293T LP cells. The fluorescence intensity of the NBN–enhanced green fluorescent protein (EGFP) fusion protein was measured by flow cytometry and normalized to the cotranslationally expressed mCherry fluorescence signal (EGFP:mCherry ratio). Each variant was tested in triplicates and protein stability was determined as the fold change to wild type (WT) NBN-EGFP expression. For western blot analysis the mCherry+BFP− population was sorted by flow cytometry. Protein samples were separated on a Mini-PROTEANTGX Precast Gel 4% to 15% (Bio-Rad Laboratories, Inc, Hercules, CA) and stained with rabbit anti-NBS1 (NB100-143; Novus Biologicals, LLC, Centennial, CO) and rabbit anti-GADPH (D16H11, Cell Signaling Technology, Inc, Danvers, MA). IRDye 800CW Goat anti-Rabbit (LI-COR Biotechnology, Lincoln, NE) was used as the secondary antibody.
For the mitomycin C (MMC) drug sensitivity assay, WT NBN and NBN variants were pooled together into a library. Next, the library was expressed in NBN−/− HEK293T LP cells and the cell pool treated with 20 nM MMC (catalog no. S8146; Selleck Chemicals LLC, Houston, TX) for 10 or 14 days. Cells were harvested for genomic DNA extraction, and Illumina MiSeq of the barcode region was performed to quantify NBN variant frequency (supplemental Table 8). Variant-barcode counts were normalized to the total barcode reads and the fold change in variant frequency after treatment compared with day 0 was used as the indicator for variant drug sensitivity. Each variant was represented by 3 barcodes and each MMC treatment condition was performed in triplicates.
Statistical analyses
The enrichment of rare and putatively damaging NBN variants in patients with B-ALL was assessed using the CoCoRV pipeline,40 in which gnomAD v.2.1.1 exome-based noncancer summary counts (n = 118 479) were used as the non-ALL control cohort. Putatively damaging NBN variants were defined as follows: allele frequency <1 × 10−3 in the general population derived from the gnomAD v2.1.1 data set,41 protein-truncating variants (frameshift and nonsense), or missense variants with a Combined Annotation Dependent Depletion score >20. Ancestry-based stratified analysis using the Cochran-Mantel-Haenszel test was used to calculate the P values. Both gene-level and specific domain-level association tests were performed.
To assess patient characteristics, we compared putatively damaging germ line NBN variant carriers (n = 47) or experimentally validated nonfunctional and partially functional germ line NBN variant carriers (n = 31), or experimentally validated nonfunctional germ line NBN variant carriers only (n = 16) with B-ALL cases with WT NBN status (n = 3719) enrolled in COG AALL0232, P9900, and St. Jude Total XIIIB and XV clinical trials. Patient characteristics included age at diagnosis, sex, genetic ancestry group, ploidy, fusion genes (ETV6::RUNX1, BCR::ABL1, TCF3::PBX1 and KMT2A-r), white blood count at diagnosis, central nervous system (CNS) status at diagnosis, end-of-induction minimal residual disease, and treatment related events. Fisher t test or nonparametric Wilcoxon rank-sum test was used to assess statistical significance. Treatment outcome (event-free survival or overall survival) was treated as a time-to-event variable and events included induction failure, relapse, and others (including second malignancies, death, and other events). Its relation to the status of germ line NBN was assessed by using the Cox regression with Firth’s penalized likelihood model adjusting for age, white blood count, and study group enrollment. We used R (v4.2.0; The R Foundation, Vienna, Austria) for all statistical analyses, unless otherwise stated.
Further details on experimental procedures and statistical analysis can be found in the supplemental Methods.
Results
Targeted sequencing of the NBN gene in pediatric B-ALL
To comprehensively characterize the pattern and prevalence of germ line NBN variants in pediatric B-ALL, we performed targeted sequencing of all exons of the NBN gene in 4325 children with newly diagnosed disease enrolled on 3 COG and 2 St. Jude frontline clinical trials (Figure 1A). In total, 77 unique NBN coding variants (including synonymous single nucleotide variant (SNV), nonsynonymous SNV, and frameshift variants) were found in B-ALL cases (supplemental Table 1). Of these, putatively damaging NBN variants were then identified based on 2 criteria: (1) a population allele frequency <1 × 10−3 in the general population derived from the gnomAD v2.1.1 data set,41 and (2) a Combined Annotation Dependent Depletion score >20.42
Workflow for NBN-targeted sequencing in pediatric patients with B-ALL. (A) CONSORT diagram of COG and St. Jude patients included in this study. (B) Protein domain plot of NBN (NM_002485): FHA, BRCT I and II, MRE11 and ATM interaction site (NBS1C), SP100 interaction site, and MTOR interaction site. The upper panel shows the amino acid substitutions predicted to result from the germ line NBN variants identified in this study. The numbers in circles indicate the number of patients that harbor the NBN variant of interest. BRCT, breast cancer C-terminus domain; FHA, forkhead-associated domain; MTOR, mechanistic target of rapamycin kinase; SNV, single nucleotide variant; SP100, SP100 nuclear antigen.
Workflow for NBN-targeted sequencing in pediatric patients with B-ALL. (A) CONSORT diagram of COG and St. Jude patients included in this study. (B) Protein domain plot of NBN (NM_002485): FHA, BRCT I and II, MRE11 and ATM interaction site (NBS1C), SP100 interaction site, and MTOR interaction site. The upper panel shows the amino acid substitutions predicted to result from the germ line NBN variants identified in this study. The numbers in circles indicate the number of patients that harbor the NBN variant of interest. BRCT, breast cancer C-terminus domain; FHA, forkhead-associated domain; MTOR, mechanistic target of rapamycin kinase; SNV, single nucleotide variant; SP100, SP100 nuclear antigen.
Overall, we discovered 25 unique and putatively damaging NBN coding variants in 50 patients with B-ALL, representing a cumulative incidence of 1.2% (Figure 1B; Table 1), and we considered these as potentially related to ALL risk. Compared with only 189 unique and putatively damaging NBN coding variants found in 472 of 118 479 noncancer individuals in the gnomAD v.2.1.1 Exomes cohort (0.4%, supplemental Figure 1; supplemental Table 2),41 putatively damaging NBN variants were significantly overrepresented in patients with B-ALL (P = .004; odds ratio [OR], 1.8; supplemental Table 3). Notably, four of 25 B-ALL–related NBN variants were not reported in the gnomAD database (Table 1). Three of the B-ALL–related NBN variants found in 9 patients resulted in protein truncation, including the known loss-of-function variant p.K219fs (Figure 1B; Table 1).11 The remaining 22 variants were missense and preferentially located in the N-terminal FHA-BRCT-repeat domain (16/22 variants, 72.7%; Figure 1B). The allele fraction of each variant in each sample was confirmed to be ∼50%, consistent with a heterozygous genotype in all carriers of putatively damaging NBN variants (supplemental Table 1).
Rare and putatively damaging germ line NBN variants identified in patients with B-ALL (NBN NM_002485)
| Variant ID . | Exon . | Class . | Genomic location . | Sequence variant . | AF . | CADD . | REVEL . | Designation∗ . | Cases . | Genetic ancestry group (cases) . |
|---|---|---|---|---|---|---|---|---|---|---|
| p.L4P | 1 | Missense | 8-90996779-A-G | c.T11C | 0 | 24.6 | 0.462 | VUS | 2 | AMR (2) |
| p.L18I | 2 | Missense | 8-90995069-G-T | c.C52A | 1.41E−05 | 24 | 0.158 | VUS | 1 | AFR (1) |
| p.Q39K | 2 | Missense | 8-90995006-G-T | c.C115A | 0 | 24 | 0.542 | VUS | 1 | AMR (1) |
| p.S40L | 2 | Missense | 8-90995002-G-A | c.C119T | 1.19E−05 | 27 | 0.88 | VUS | 2 | EUR (2) |
| p.S93L | 3 | Missense | 8-90993645-G-A | c.C278T | 5.81E−04 | 25 | 0.457 | VUS† | 2 | EUR (2) |
| p.M152I | 4 | Missense | 8-90992986-C-T | c.G456A | 1.13E−04 | 27.3 | 0.482 | VUS† | 7 | EUR (4), AMR (2), Other (1) |
| p.K156N | 4 | Missense | 8-90992974-T-G | c.A468C | 1.51E−04 | 26.1 | 0.438 | VUS† | 3 | AMR (3) |
| p.E179K | 5 | Missense | 8-90990497-C-T | c.G535A | 3.98E−06 | 32 | 0.308 | VUS | 1 | EUR (1) |
| p.V184A | 5 | Missense | 8-90990481-A-G | c.T551C | 0 | 25.2 | 0.179 | VUS | 1 | Other (1) |
| p.E217Q | 6 | Missense | 8-90983454-C-G | c.G649C | 7.97E−06 | 25.1 | 0.224 | VUS | 1 | EUR (1) |
| p.K219fs | 6 | Frameshift | 8-90983441-ATTTGT-A | c.657_661del | 2.02E−04 | 32 | NA | Pathogenic | 7 | EUR (5), AMR (2) |
| p.F222L | 6 | Missense | 8-90983439-A-G | c.T664C | 1.77E−05 | 28.2 | 0.937 | VUS | 1 | EUR (1) |
| p.I228R | 6 | Missense | 8-90983420-A-C | c.T683G | 7.10E−05 | 25.7 | 0.54 | VUS | 3 | EUR (2), AFR (1) |
| p.A241T | 7 | Missense | 8-90982767-C-T | c.G721A | 1.59E−05 | 26 | 0.396 | VUS | 1 | AMR (1) |
| p.F263S | 7 | Missense | 8-90982700-A-G | c.T788C | 1.95E−04 | 25.4 | 0.737 | VUS† | 2 | AFR (1), Other (1) |
| p.G274R | 7 | Missense | 8-90982668-C-T | c.G820A | 7.96E−06 | 24 | 0.308 | VUS | 1 | AFR (1) |
| p.L281X | 7 | Stop gain | 8-90982646-A-C | c.T842G | 3.98E−06 | 33 | NA | Pathogenic | 1 | AMR (1) |
| p.A313V | 8 | Missense | 8-90976694-G-A | c.C938T | 3.18E−05 | 26.8 | 0.661 | VUS | 1 | AMR (1) |
| p.Q448L | 10 | Missense | 8-90967565-T-A | c.A1343T | 4.25E−05 | 21.5 | 0.037 | VUS† | 1 | AFR (1) |
| p.D469Y | 11 | Missense | 8-90965912-C-A | c.G1405T | 6.16E−05 | 26.1 | 0.149 | VUS† | 1 | AFR (1) |
| p.P495L | 11 | Missense | 8-90965833-G-A | c.C1484T | 3.19E−05 | 23.9 | 0.097 | VUS | 1 | EUR (1) |
| p.E552Q | 11 | Missense | 8-90965663-C-G | c.G1654C | 0 | 23.6 | 0.058 | VUS | 1 | EUR (1) |
| p.V556E | 11 | Missense | 8-90965650-A-T | c.T1667A | 1.99E−05 | 22.5 | 0.078 | VUS | 2 | EUR (2) |
| p.E564K | 11 | Missense | 8-90965627-C-T | c.G1690A | 8.10E−04 | 22.9 | 0.081 | (Likely) Benign | 5 | EUR (1), EAS (2), SAS (1), Other (1) |
| p.S706X | 14 | Stop gain | 8-90955548-G-C | c.C2117G | 1.06E−05 | 42 | NA | Pathogenic | 1 | EUR (1) |
| Variant ID . | Exon . | Class . | Genomic location . | Sequence variant . | AF . | CADD . | REVEL . | Designation∗ . | Cases . | Genetic ancestry group (cases) . |
|---|---|---|---|---|---|---|---|---|---|---|
| p.L4P | 1 | Missense | 8-90996779-A-G | c.T11C | 0 | 24.6 | 0.462 | VUS | 2 | AMR (2) |
| p.L18I | 2 | Missense | 8-90995069-G-T | c.C52A | 1.41E−05 | 24 | 0.158 | VUS | 1 | AFR (1) |
| p.Q39K | 2 | Missense | 8-90995006-G-T | c.C115A | 0 | 24 | 0.542 | VUS | 1 | AMR (1) |
| p.S40L | 2 | Missense | 8-90995002-G-A | c.C119T | 1.19E−05 | 27 | 0.88 | VUS | 2 | EUR (2) |
| p.S93L | 3 | Missense | 8-90993645-G-A | c.C278T | 5.81E−04 | 25 | 0.457 | VUS† | 2 | EUR (2) |
| p.M152I | 4 | Missense | 8-90992986-C-T | c.G456A | 1.13E−04 | 27.3 | 0.482 | VUS† | 7 | EUR (4), AMR (2), Other (1) |
| p.K156N | 4 | Missense | 8-90992974-T-G | c.A468C | 1.51E−04 | 26.1 | 0.438 | VUS† | 3 | AMR (3) |
| p.E179K | 5 | Missense | 8-90990497-C-T | c.G535A | 3.98E−06 | 32 | 0.308 | VUS | 1 | EUR (1) |
| p.V184A | 5 | Missense | 8-90990481-A-G | c.T551C | 0 | 25.2 | 0.179 | VUS | 1 | Other (1) |
| p.E217Q | 6 | Missense | 8-90983454-C-G | c.G649C | 7.97E−06 | 25.1 | 0.224 | VUS | 1 | EUR (1) |
| p.K219fs | 6 | Frameshift | 8-90983441-ATTTGT-A | c.657_661del | 2.02E−04 | 32 | NA | Pathogenic | 7 | EUR (5), AMR (2) |
| p.F222L | 6 | Missense | 8-90983439-A-G | c.T664C | 1.77E−05 | 28.2 | 0.937 | VUS | 1 | EUR (1) |
| p.I228R | 6 | Missense | 8-90983420-A-C | c.T683G | 7.10E−05 | 25.7 | 0.54 | VUS | 3 | EUR (2), AFR (1) |
| p.A241T | 7 | Missense | 8-90982767-C-T | c.G721A | 1.59E−05 | 26 | 0.396 | VUS | 1 | AMR (1) |
| p.F263S | 7 | Missense | 8-90982700-A-G | c.T788C | 1.95E−04 | 25.4 | 0.737 | VUS† | 2 | AFR (1), Other (1) |
| p.G274R | 7 | Missense | 8-90982668-C-T | c.G820A | 7.96E−06 | 24 | 0.308 | VUS | 1 | AFR (1) |
| p.L281X | 7 | Stop gain | 8-90982646-A-C | c.T842G | 3.98E−06 | 33 | NA | Pathogenic | 1 | AMR (1) |
| p.A313V | 8 | Missense | 8-90976694-G-A | c.C938T | 3.18E−05 | 26.8 | 0.661 | VUS | 1 | AMR (1) |
| p.Q448L | 10 | Missense | 8-90967565-T-A | c.A1343T | 4.25E−05 | 21.5 | 0.037 | VUS† | 1 | AFR (1) |
| p.D469Y | 11 | Missense | 8-90965912-C-A | c.G1405T | 6.16E−05 | 26.1 | 0.149 | VUS† | 1 | AFR (1) |
| p.P495L | 11 | Missense | 8-90965833-G-A | c.C1484T | 3.19E−05 | 23.9 | 0.097 | VUS | 1 | EUR (1) |
| p.E552Q | 11 | Missense | 8-90965663-C-G | c.G1654C | 0 | 23.6 | 0.058 | VUS | 1 | EUR (1) |
| p.V556E | 11 | Missense | 8-90965650-A-T | c.T1667A | 1.99E−05 | 22.5 | 0.078 | VUS | 2 | EUR (2) |
| p.E564K | 11 | Missense | 8-90965627-C-T | c.G1690A | 8.10E−04 | 22.9 | 0.081 | (Likely) Benign | 5 | EUR (1), EAS (2), SAS (1), Other (1) |
| p.S706X | 14 | Stop gain | 8-90955548-G-C | c.C2117G | 1.06E−05 | 42 | NA | Pathogenic | 1 | EUR (1) |
AF, allele frequency in gnomAD; AFR, African/African American; AMR, admixed American; CADD, Combined Annotation Dependent Depletion; EAS, East Asian; EUR, European (non-Finnish); SAS, South Asian; VUS, variant of uncertain significance.
NBN variant designation reported in the ClinVar database (accession date: March 2023).
VUS with conflicting interpretations of pathogenicity.
Functional characterization of NBN variants
Of the 25 B-ALL–related NBN variants in our cohort, only 4 variants (16%) are classified as benign or pathogenic as reported in the ClinVar database, with the remaining 21 variants (84%) noted as “of uncertain significance” (Table 1). To comprehensively characterize B-ALL–related germ line NBN variants, we utilized the HEK293T LP model to examine variant function at a single cell level.37,38 First, we knocked out the endogenous NBN gene by CRISPR/Cas9 editing (hereafter, NBN−/− HEK293T LP cells). Next, we generated the 25 NBN variants of interest by site-directed mutagenesis, with each variant tagged with an EGFP-fusion protein and a unique barcode index.39NBN variants were introduced into the attP site of NBN−/− HEK293T LP cells by homology recombination such that each cell expresses only a single variant of interest. Finally, NBN variants were subjected to 2 phenotyping assays to determine their effect on (1) NBN protein stability and (2) MMC drug sensitivity in vitro (Figure 2).
Experimental design for NBN variant functional characterization. Parallel NBN variant characterization was done using the engineered NBN−/− HEK293T LP cell line model. First, the 25 variants of interest were fused to EGPF, tagged with a unique barcode sequence, and cloned in the attB-mCherry recombination plasmid. Next, NBN variants were introduced into the attP/attB recombination site in NBN−/− HEK293T LP cells. Cells with successful recombination were identified as mCherry+/BFP− population in flow cytometry. Finally, NBN variant expressing cells were subjected to 2 different types of phenotyping to determine their effect on (1) NBN variant protein stability or (2) NBN variant MMC sensitivity in vitro. NBN protein stability was quantified by the fluorescence intensity of the EGFP-fusion protein and normalized to the co-translationally expressed mCherry fluorescence signal (EGFP:mCherry ratio). Unstable variants resulted in decreased EGFP expression and thus a low EGFP:mCherry ratio, as illustrated by the red-colored histogram. Drug sensitivity was determined by the change in NBN variant frequency after MMC exposure. Damaging variants resulted in the loss of NBN signaling during MMC-induced DNA-damage repair. This led to reduced cell survival and thus under-representation of the respective variants after MMC treatment, which was quantified by targeted sequencing of the barcode region. Finally, protein stability and MMC drug sensitivity were both considered for NBN variant classification. BFP, blue fluorescent protein; EGPF, enhanced green fluorescent protein; MMC, mitomycin C.
Experimental design for NBN variant functional characterization. Parallel NBN variant characterization was done using the engineered NBN−/− HEK293T LP cell line model. First, the 25 variants of interest were fused to EGPF, tagged with a unique barcode sequence, and cloned in the attB-mCherry recombination plasmid. Next, NBN variants were introduced into the attP/attB recombination site in NBN−/− HEK293T LP cells. Cells with successful recombination were identified as mCherry+/BFP− population in flow cytometry. Finally, NBN variant expressing cells were subjected to 2 different types of phenotyping to determine their effect on (1) NBN variant protein stability or (2) NBN variant MMC sensitivity in vitro. NBN protein stability was quantified by the fluorescence intensity of the EGFP-fusion protein and normalized to the co-translationally expressed mCherry fluorescence signal (EGFP:mCherry ratio). Unstable variants resulted in decreased EGFP expression and thus a low EGFP:mCherry ratio, as illustrated by the red-colored histogram. Drug sensitivity was determined by the change in NBN variant frequency after MMC exposure. Damaging variants resulted in the loss of NBN signaling during MMC-induced DNA-damage repair. This led to reduced cell survival and thus under-representation of the respective variants after MMC treatment, which was quantified by targeted sequencing of the barcode region. Finally, protein stability and MMC drug sensitivity were both considered for NBN variant classification. BFP, blue fluorescent protein; EGPF, enhanced green fluorescent protein; MMC, mitomycin C.
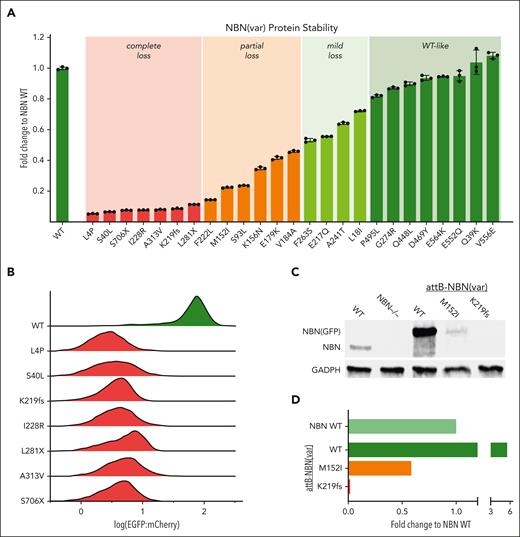
We focused on protein stability because this is a known determinant of NBN activity. In addition, in patients with NBS with p.K219fs genotype, the expression level of the alternatively translated protein, p70, significantly correlates with cancer risk.43 We determined protein stability for each NBN variant by measuring the fluorescence intensity of the EGFP-fusion protein as a proxy marker for the variant protein abundance (Figure 2).37,39 As shown in Figure 3A-B, WT NBN tagged with EGFP resulted in robust green fluorescence as measured with flow cytometry. By contrast, the loss-of-function NBN variant, p.K219fs, led to an ∼10-fold reduction in the EGFP signal. Applying this assay to all B-ALL–related NBN variants, we identified another 6 variants that resulted in unstable protein, defined as ≥90% reduction of EGFP intensity compared with WT NBN-EGFP (Figure 3A). As expected, all 3 of the truncating variants resulted in loss of protein expression. However, 4 of the unstable variants are missense, leading to single amino acid substitutions at positions 4, 40, 228, and 313 and resulting in complete loss of protein stability as well (Figure 3A-B). Six missense variants showed partial loss of NBN protein stability (defined as 50%-90% reduction of EGFP intensity); 4 variants showed only mild loss (20%-50% reduction of EGFP intensity); and 8 variants had WT-like protein stability (<20% reduction of EGFP intensity). To validate these results using orthogonal assays, we selected the most common missense variant, p.M152I, and the most common truncating variant, p.K219fs, to perform western blot analysis (Figure 3C-D).
NBN variant protein stability screen. (A) Presentation of the fold change of NBN variant EGFP:mCherry ratio compared with WT NBN as quantified by flow cytometry: complete loss <0.1, partial loss 0.1 to 0.5, mild loss 0.5 to 0.8, WT-like >0.8. The results are presented as the mean of 3 independent experiments ± standard deviation. (B) The pattern of EGFP:mCherry distribution in unstable NBN variants (red) compared with WT NBN (green). Each histogram was generated from ∼4000 NBN (WT or variant) expressing cells. (C) Western blot analysis of NBN (WT or variant) protein expression level. Lanes 1 to 2 show the absence of WT NBN protein expression in NBN−/− HEK293T LP cells (lane 2) compared with WT HEK293T LP cells (lane 1). Lanes 3 to 5 depict NBN (WT or variant) protein levels after reexpression in NBN−/− HEK293T LP cells. Successfully recombined cells were identified as mCherry+/BFP− population and separated by flow cytometry before protein extraction and western blot analysis. The EGFP-fusion protein resulted in a slight increase in the molecular weight. (D) Relative NBN variant expression level compared with WT NBN expression as quantified using western blot.
NBN variant protein stability screen. (A) Presentation of the fold change of NBN variant EGFP:mCherry ratio compared with WT NBN as quantified by flow cytometry: complete loss <0.1, partial loss 0.1 to 0.5, mild loss 0.5 to 0.8, WT-like >0.8. The results are presented as the mean of 3 independent experiments ± standard deviation. (B) The pattern of EGFP:mCherry distribution in unstable NBN variants (red) compared with WT NBN (green). Each histogram was generated from ∼4000 NBN (WT or variant) expressing cells. (C) Western blot analysis of NBN (WT or variant) protein expression level. Lanes 1 to 2 show the absence of WT NBN protein expression in NBN−/− HEK293T LP cells (lane 2) compared with WT HEK293T LP cells (lane 1). Lanes 3 to 5 depict NBN (WT or variant) protein levels after reexpression in NBN−/− HEK293T LP cells. Successfully recombined cells were identified as mCherry+/BFP− population and separated by flow cytometry before protein extraction and western blot analysis. The EGFP-fusion protein resulted in a slight increase in the molecular weight. (D) Relative NBN variant expression level compared with WT NBN expression as quantified using western blot.
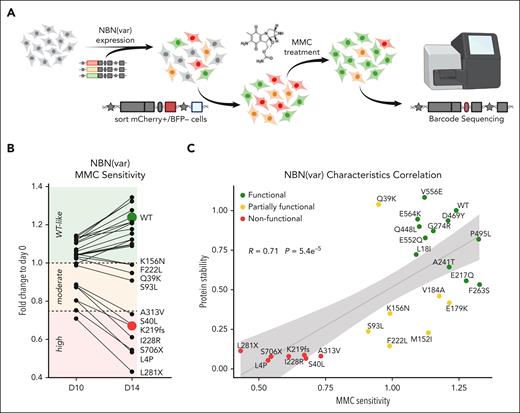
Because protein instability is not the only mechanism for loss of activity, we sought to evaluate a cellular end point that more broadly reflects NBN activity. Cells derived from patients with NBS (eg, fibroblasts or lymphoblastoid cells) show increased chromosomal aberrations and decreased proliferation after DNA damage induced by irradiation or exposure to radiomimetic drugs such as MMC.8,44,45 In fact, MMC sensitivity testing is an established clinical assay for the diagnosis of NBS. To demonstrate the validity of this assay, we first confirmed that NBN−/− HEK293T LP cells are highly sensitive to MMC and reexpression of WT NBN greatly enhanced tolerance to MMC-induced apoptosis (supplemental Figure 2A-B). By contrast, reexpression of the known pathogenic variant, p.K219fs, conferred no survival benefit during MMC treatment. Next, WT NBN and 25 NBN variants of interest were pooled into a library and were expressed in NBN−/− HEK293T LP cells. Because cells expressing a loss-of-function NBN variant are more susceptible to MMC-induced DNA damage, these cells are expected to undergo apoptosis quickly and the respective variants should become underrepresented after MMC exposure (Figure 4A). Based on the fold change in variant frequency at day 14 relative to pre-MMC treatment, we assigned each variant as “highly sensitive” (fold change, <0.75), “moderately sensitive” (fold change, 0.75-1), or “WT-like” (fold change, >1), respectively (Figure 4B). Besides the known loss-of-function variant, p.K219fs, 2 truncating variants (p.L281X and p.S706X) and 4 missense variants (p.L4P, p.S40L, p.I228R, and p.A313V) showed poor survival during MMC treatment and were thus designated as highly sensitive. In addition, 4 variants (p.Q39K, p.S93L, p.K156N, and p.F222L) were tested as moderately sensitive to MMC.
NBN variant MMC drug sensitivity screen. (A) WT NBN or 25 NBN variants were expressed in NBN−/− HEK293T LP cells. Successfully recombined cells were identified as mCherry+/BFP− population, separated by flow cytometry, and cultured in media supplemented with MMC 20 nM for 10 or 14 days. NBN−/− HEK293T LP cells expressing WT NBN or WT-like variants (green) became more tolerant to MMC-induced DNA damage, which resulted in higher proliferation compared with cells expressing NBN variants with reduced (orange) or loss-of-function (red) activity. MMC drug sensitivity was determined by the fold change in variant frequency before and after MMC exposure and was quantified by Illumina MiSeq of the barcode sequence for each variant. (B) NBN variant abundance during 20 nM MMC treatment for 10 and 14 days was measured as the fold change to day 0 of the normalized barcode reads. MMC sensitivity was classified as high (<0.75), moderate (0.75-1) and WT-like (>1). Each dot represents an average fold change of 9 measurements, which derive from 3 barcodes assigned to each NBN variant and each condition performed as triplicates. (C) NBN variant protein stability was plotted against NBN variant MMC sensitivity. P values were estimated using the Pearson correlation test (R).
NBN variant MMC drug sensitivity screen. (A) WT NBN or 25 NBN variants were expressed in NBN−/− HEK293T LP cells. Successfully recombined cells were identified as mCherry+/BFP− population, separated by flow cytometry, and cultured in media supplemented with MMC 20 nM for 10 or 14 days. NBN−/− HEK293T LP cells expressing WT NBN or WT-like variants (green) became more tolerant to MMC-induced DNA damage, which resulted in higher proliferation compared with cells expressing NBN variants with reduced (orange) or loss-of-function (red) activity. MMC drug sensitivity was determined by the fold change in variant frequency before and after MMC exposure and was quantified by Illumina MiSeq of the barcode sequence for each variant. (B) NBN variant abundance during 20 nM MMC treatment for 10 and 14 days was measured as the fold change to day 0 of the normalized barcode reads. MMC sensitivity was classified as high (<0.75), moderate (0.75-1) and WT-like (>1). Each dot represents an average fold change of 9 measurements, which derive from 3 barcodes assigned to each NBN variant and each condition performed as triplicates. (C) NBN variant protein stability was plotted against NBN variant MMC sensitivity. P values were estimated using the Pearson correlation test (R).
NBN variant classification based on protein stability and MMC sensitivity
Based on functional characterization results, we classified the 25 B-ALL–related NBN variants as “nonfunctional,” “partially functional,” and “functional,” as summarized in Table 2. Comparing the results from both screening approaches, we found a strong correlation between NBN variant protein stability and MMC drug sensitivity (r = 0.71, P = 5.4 × 10−5, Pearson correlation test, Figure 4C). Besides p.K219fs, 6 variants were linked to both, unstable protein and high sensitivity to MMC, and thus designated as nonfunctional (Figure 4C; Table 2). Further, 11 variants were found to have WT-like MMC tolerance and WT-like or mild loss of NBN protein stability and were therefore assigned as functional. Finally, 7 variants were considered as partially functional due to partial loss of protein stability and moderate sensitivity to MMC.
B-ALL–related NBN variant classification
| Variant . | Position . | Protein stability∗ . | MMC sensitivity† . | Final classification . |
|---|---|---|---|---|
| L4P | 4 | Complete loss | High | Nonfunctional |
| S40L | 40 | Complete loss | High | Nonfunctional |
| S706X | 706 | Complete loss | High | Nonfunctional |
| I228R | 228 | Complete loss | High | Nonfunctional |
| A313V | 313 | Complete loss | High | Nonfunctional |
| K219fs | 219 | Complete loss | High | Nonfunctional |
| L281X | 281 | Complete loss | High | Nonfunctional |
| F222L | 222 | Partial loss | Moderate | Partially functional |
| M152I | 152 | Partial loss | WT-like | Partially functional |
| S93L | 93 | Partial loss | Moderate | Partially functional |
| K156N | 156 | Partial loss | Moderate | Partially functional |
| E179K | 179 | Partial loss | WT-like | Partially functional |
| V184A | 184 | Partial loss | WT-like | Partially functional |
| Q39K | 39 | WT-like | Moderate | Partially functional |
| F263S | 263 | Mild loss | WT-like | Functional |
| E217Q | 217 | Mild loss | WT-like | Functional |
| A241T | 241 | Mild loss | WT-like | Functional |
| L18I | 18 | Mild loss | WT-like | Functional |
| P495L | 495 | WT-like | WT-like | Functional |
| E552Q | 552 | WT-like | WT-like | Functional |
| G274R | 274 | WT-like | WT-like | Functional |
| Q448L | 448 | WT-like | WT-like | Functional |
| D469Y | 469 | WT-like | WT-like | Functional |
| E564K | 564 | WT-like | WT-like | Functional |
| V556E | 556 | WT-like | WT-like | Functional |
| Variant . | Position . | Protein stability∗ . | MMC sensitivity† . | Final classification . |
|---|---|---|---|---|
| L4P | 4 | Complete loss | High | Nonfunctional |
| S40L | 40 | Complete loss | High | Nonfunctional |
| S706X | 706 | Complete loss | High | Nonfunctional |
| I228R | 228 | Complete loss | High | Nonfunctional |
| A313V | 313 | Complete loss | High | Nonfunctional |
| K219fs | 219 | Complete loss | High | Nonfunctional |
| L281X | 281 | Complete loss | High | Nonfunctional |
| F222L | 222 | Partial loss | Moderate | Partially functional |
| M152I | 152 | Partial loss | WT-like | Partially functional |
| S93L | 93 | Partial loss | Moderate | Partially functional |
| K156N | 156 | Partial loss | Moderate | Partially functional |
| E179K | 179 | Partial loss | WT-like | Partially functional |
| V184A | 184 | Partial loss | WT-like | Partially functional |
| Q39K | 39 | WT-like | Moderate | Partially functional |
| F263S | 263 | Mild loss | WT-like | Functional |
| E217Q | 217 | Mild loss | WT-like | Functional |
| A241T | 241 | Mild loss | WT-like | Functional |
| L18I | 18 | Mild loss | WT-like | Functional |
| P495L | 495 | WT-like | WT-like | Functional |
| E552Q | 552 | WT-like | WT-like | Functional |
| G274R | 274 | WT-like | WT-like | Functional |
| Q448L | 448 | WT-like | WT-like | Functional |
| D469Y | 469 | WT-like | WT-like | Functional |
| E564K | 564 | WT-like | WT-like | Functional |
| V556E | 556 | WT-like | WT-like | Functional |
Variant protein stability compared with WT NBN stability: complete loss ≥90% reduction, partial loss 50% to 90% reduction, mild loss 20% to 50% reduction, WT-like <20% reduction.
MMC sensitivity quantified as fold change in variant frequency pre- and post-MMC treatment: high <0.75, moderate 0.75 to 1, WT-like >1.
Nonfunctional NBN variants in B-ALL cases cluster in the N-terminal functional domains
Of the 14 NBN variants experimentally validated as nonfunctional or partially functional (Table 2), 13 affect the N-terminal FHA-BRCT-repeat domain (Figure 5A). Sequence alignment confirmed this region to be highly conserved among different species and AlphaFold prediction identified the FHA-BRCT–repeat domain as a complex convoluted tertiary structure (Figure 5B).46 Therefore, we reasoned that sequence variation of the FHA-BRCT–repeat domain may have strong effects on NBN function. In fact, variants within the N-terminus domain showed a heightened association with B-ALL risk (P = .003; OR, 2) and remained significant even after excluding p.K219fs from the analysis (P = .02; OR, 1.8; Figure 5C; supplemental Table 3).
Key functional domains of NBN are preferentially affected by genetic variation. (A) The top panel summarizes the frequency in B-ALL cases, effects on MMC drug sensitivity, and NBN protein stability for each NBN variant. The bottom panel depicts the alignment of NBN protein sequences from human (Homo sapiens; NP_002476.2), mouse (Mus musculus, NP_038780.3), rat (Rattus norvegicus; NP_620228.1), and monkey (Macaca mulatta; NP_001252668.1). Protein sequence alignment was done in COBALT NCBI Multiple Sequence Alignment Viewer, version 1.22.0. (B) AlphaFold structure prediction of NIBRIN (AF-O60934-F1): FAH domain in orange, BRCT I and II in red, and C-terminal domain in green. Experimentally validated functional NBN variants relate to the peripheral moieties, whereas nonfunctional and partially functional NBN variants relate to the NBN central region. (C) Cumulative burden of putatively damaging NBN variants in B-ALL cases vs gnomAD noncancer controls calculated by ancestry-based stratified analysis using the Cochran-Mantel-Haenszel test. ∗P < .05 and ∗∗P < .01.
Key functional domains of NBN are preferentially affected by genetic variation. (A) The top panel summarizes the frequency in B-ALL cases, effects on MMC drug sensitivity, and NBN protein stability for each NBN variant. The bottom panel depicts the alignment of NBN protein sequences from human (Homo sapiens; NP_002476.2), mouse (Mus musculus, NP_038780.3), rat (Rattus norvegicus; NP_620228.1), and monkey (Macaca mulatta; NP_001252668.1). Protein sequence alignment was done in COBALT NCBI Multiple Sequence Alignment Viewer, version 1.22.0. (B) AlphaFold structure prediction of NIBRIN (AF-O60934-F1): FAH domain in orange, BRCT I and II in red, and C-terminal domain in green. Experimentally validated functional NBN variants relate to the peripheral moieties, whereas nonfunctional and partially functional NBN variants relate to the NBN central region. (C) Cumulative burden of putatively damaging NBN variants in B-ALL cases vs gnomAD noncancer controls calculated by ancestry-based stratified analysis using the Cochran-Mantel-Haenszel test. ∗P < .05 and ∗∗P < .01.
Patient characteristics and outcome analysis for germ line NBN variant carriers
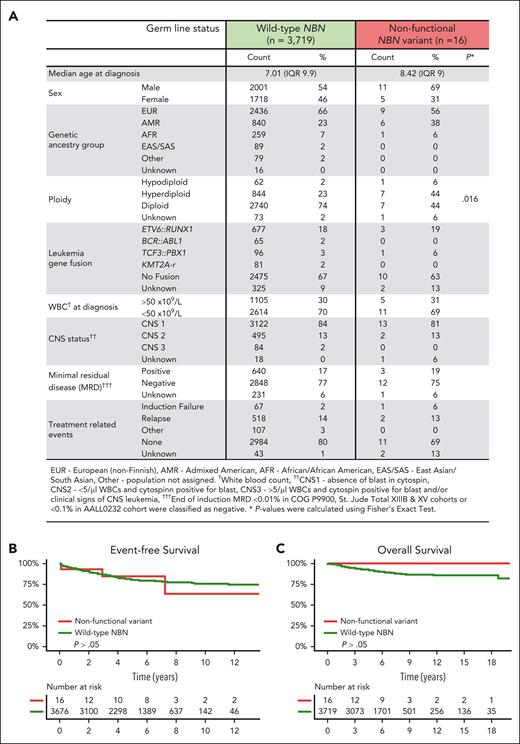
Next, we examined the relationship between germ line NBN status and clinical features of B-ALL in 4 large clinical trials (COG P9900, AALL0232, St. Jude Total XIIIB, and Total XV). Comparison of 47 NBN variant carriers and 3719 patients with WT NBN status did not reveal a significant difference in patient characteristics (age at diagnosis, sex, or genetic ancestry group) or leukemia genetic subtype (ploidy and fusion genes). Restricting this analysis to patients with experimentally validated nonfunctional and partially functional NBN variants (n = 31) did not show any significant association either (supplemental Figure 3A). Finally, focusing on cases with validated nonfunctional variants only showed an enrichment of hyperdiploid B-ALL in this small cohort of 16 carriers (Figure 6A). The NBS founder mutation, p.K219fs, is most frequently found in Eastern European populations; in fact, 4 of 7 carriers of this variant in our ALL cohort were of European descent (Table 1).12,47 However, among all NBN variant carriers, there did not seem to be an enrichment of European ancestry (Figure 6A). In fact, B-ALL–related NBN variants were also frequently found in the admixed American population.
Association of NBN variants with clinical characteristics of B-ALL. (A) Characteristics of B-ALL in carriers of an experimentally validated nonfunctional germ line NBN variant (n = 16) were compared with those with WT NBN status (n = 3719) treated in COG P9900, AALL0232, St. Jude Total XIIIB and St. Jude Total XV clinical trials. (B) Event-free survival and (C) overall survival in carriers of an experimentally validated nonfunctional germ line NBN variant and patients with WT NBN status.
Association of NBN variants with clinical characteristics of B-ALL. (A) Characteristics of B-ALL in carriers of an experimentally validated nonfunctional germ line NBN variant (n = 16) were compared with those with WT NBN status (n = 3719) treated in COG P9900, AALL0232, St. Jude Total XIIIB and St. Jude Total XV clinical trials. (B) Event-free survival and (C) overall survival in carriers of an experimentally validated nonfunctional germ line NBN variant and patients with WT NBN status.
Finally, we evaluated the association of germ line NBN status and treatment outcomes of patients treated on 2 COG and 2 St. Jude frontline B-ALL clinical trials (AALL0232, COG P9900, St. Jude Total XIIIB and XV). Carriers of nonfunctional NBN variants showed similar response to induction chemotherapy compared with those with WT NBN status, as measured by the minimal residual disease (Figure 6A). Unlike NBS patients, who have been reported to have an inferior prognosis,48 there were also no significant differences in overall survival or event-free survival between heterozygous carriers of nonfunctional NBN variants and those with WT NBN status in our cohort (Figure 6B-C; supplemental Figure 3B-C). Notably, no nonfunctional NBN variant carrier was diagnosed with a second malignancy during a median follow-up period of 6.15 years (range, 1.1 month-13.7 years).
Discussion
Even though carriers of a heterozygous germ line NBN variant have been linked to cancer risk,20,22,23,49,50 malignancies of lymphatic origin, in particular B-ALL, were underrepresented in these studies.21 Therefore, evaluation of this patient group has been limited. Herein, we comprehensively characterized NBN-related genetic predisposition to pediatric B-ALL, enabled by sequencing 4325 leukemia samples and systematic functional characterization of the risk variants. The large sample size of our cohort enabled the identification of extremely rare but highly deleterious variants, many of which (13/25, 52%) were described for the first time in cancer (Catalogue of Somatic Mutations in Cancer [COSMIC] v98, accession date 31 October 2023). Based on case-control variant burden analysis, we identified significant overrepresentation of putatively damaging NBN variants, most of which are missense. Moreover, when NBN variants were evaluated individually, several showed significant enrichment in B-ALL cases compared with gnomAD controls, including the NBS founder mutation, p.K219fs (supplemental Table 4). By utilizing the LP system, we established a cellular screening platform to characterize NBN variant functional properties at-scale, identifying 14 as nonfunctional or partially functional. Outcome analyses in 4 frontline ALL clinical trials at COG and St. Jude suggest that heterozygous carriers of nonfunctional NBN variants fare as well as those with WT NBN status. Overall, our study advanced the understanding of the role of germ line NBN variants in pediatric B-ALL predisposition and treatment.
B-ALL cases for NBN-targeted sequencing were chosen mostly based on sample availability from frontline ALL clinical trials, although sampling bias cannot be completely ruled out. Case-control variant burden analysis was done in CoCoRV,40 a framework specifically developed for using public summary counts as controls. The pipeline includes several features to account for potential confounders, especially population structure, which is addressed by ancestry-based stratified analysis using the Cochran-Mantel-Haenszel test. In fact, we repeated our analysis using only synonymous NBN variants and found them to be equally frequent in cases and controls (P = 1; OR, 0.99), suggesting our analytical approach was unlikely to produce spurious associations. Nonetheless, future efforts to sequence large numbers of ALL cases and controls, matched particularly for population structure and ancestry, are needed to more accurately estimate the impact (effect size) of pathogenic NBN variants on ALL risk.
The majority of known pathogenic NBN variants lead to protein truncation, resulting in partially functional protein.21,51 These hypomorphic variants retain some activity in DNA-damage response and variability in their expression level seems to be linked to the degree of genome instability and cancer risk.43 This observation is supported by our finding that NBN protein stability and MMC drug sensitivity are strongly correlated. In addition, missense NBN variants may severely impair NBN signaling, as has been suggested in a single patient with NBS with compound heterozygous genotype for p.K219fs and p.R215W and particularly severe NBS phenotype.52,53 In patients with B-ALL, we detected missense variants with impaired function far more frequently than truncating variants, and we found missense variants to be enriched in the FHA-BRCT-repeat domain. The pathogenicity of these variants may be explained by the moderate to severe reduction in protein stability. However, these variants within the highly conserved FHA-BRCT-repeat domain may impair its ability to protein interaction and to recruit the MRE11-RAD50-NBS1 (MRN) complex.13,14,54 Consistent with this hypothesis, one partially functional missense variant, p.Q39K, did not affect protein stability. This variant alters the FHA phosphoprotein-binding pocket that interacts with the endonuclease CtIP.54 Sequence variation at adjacent positions were shown to sensitize to irradiation or camptothecin in vitro but without affecting NBN stability, similar to what we observe for the NBN variant p.Q39K.54
NBN-deficient cells display elevated levels of baseline DNA damage, chromosomal instability, and aberrant cell cycle control,55 which is considered to drive cancer development. In lymphatic tissues, loss of NBN expression results in profound defects in the hematopoietic stem cell and lymphoid differentiation process.55-58 This phenotype is attributed to the impaired resolution of recombination activating gene induced DNA double-strand breaks during V(D)J recombination,58 which may also explain the particularly high risk for leukemia and lymphoma development. Regrettably, we lack detailed immunophenotyping data to inform the lymphoid compartment in germ line NBN variant carriers in our cohort. Furthermore, our cohort was limited to B-ALL cases, although T-lineage lymphoid malignancies are also frequently seen in patients with NBS.59
Using the LP system, we investigated each variant under identical experimental conditions and with the same genetic background. This allowed us to compare NBN activity across variants, using WT NBN as the reference. However, this technique is limited to in vitro characterization and cannot recapitulate NBN variant mediated leukemogenesis, which would require in vivo techniques. Therefore, the molecular mechanism of B-ALL development in these individuals remains to be investigated.
Despite the high incidence of cancer in patients with NBS, no NBN variant carrier in our cohort was diagnosed with a second malignancy, which may have been confounded by the length of follow-up. Family history was available based on the patient interview for 6 cases enrolled on St. Jude clinical trials, of whom 5 have an unremarkable history. Only 1 case, carrier of the partially functional NBN variant p.M152I and with low-hypodiploid B-ALL, showed multiple occurrences of cancer in family history. However, this individual also has a germ line variant in the TP53 gene (rs1042522), which is well recognized to be associated with low-hypodiploid ALL.60 Therefore, it is unclear to what degree the NBN variant contributed to the familial tumorigenesis in this patient.
In NBN-deficient mice with T-cell lymphoma, tumor genomic analysis revealed a characteristic mutational pattern.56 For 2 patients from the St. Jude Total XV cohort, we were able to examine somatic single nucleotide alterations using matched tumor/germ line whole-genome sequencing and compared with single base substitutions signatures in COSMIC Mutational Signatures version 3.3, which was found to be unremarkable. Further we focused on large structural alterations but did not observe any deletions affecting the NBN locus. A limited number of 4 cases had tumor whole-exome sequencing data available and we ruled out a second somatic mutation or indels affecting the NBN locus. These findings suggest the need for future studies exploring the impact of heterozygous NBN variants on lymphoid differentiation and leukemia development.
Taken together, our results highlight the importance of germ line pathogenic NBN variants to B-ALL predisposition, which may inform clinical strategies and cancer surveillance in these children in the future.
Acknowledgments
This work was supported by the National Institutes of Health (NIH), National Institute of General Medical Sciences grant P50GM115279 and NIH, National Cancer Institute (NCI) grants R01CA241452 and P30CA21765. Children’s Oncology Group clinical trials were supported by NIH/NCI grants U10 CA98543, U10 CA98413, U10 CA180886, and U10 CA180899. C.S.E. receives support through the Walter Benjamin Fellowship awarded by the German Research Foundation.
Authorship
Contribution: J.J.Y. initiated and led the project; C.-H.P., S.P.H., M.L.L., and J.J.Y. designed the study; E.A.R., M.D., K.E.N., H.I., C.-H.P., S.J., B.M.C., E.L., S.P.H., and M.L.L. contributed to the data collection; C.S.E., W.C., G.W., W.Y., and Z.L. analyzed genomic and patient data; C.S.E., Y.L., and R.N. designed functional experiments; C.S.E. performed functional experiments; C.S.E. and J.J.Y. interpreted the data and wrote the manuscript; and all authors reviewed and approved the manuscript.
Conflict-of-interest disclosure: J.J.Y. receives research funding from Takeda Pharmaceutical and AstraZeneca. S.P.H. holds the Jeffrey E. Perelman distinguished Chair in Pediatrics at the Children’s Hospital of Philadelphia. None of these relationships were related to this work. The remaining authors declare no competing financial interests.
Correspondence: Jun J. Yang, St. Jude Children's Research Hospital, Department of Pharmacy and Pharmaceutical Sciences, 262 Danny Thomas Place, Memphis, TN 38105; email: jun.yang@stjude.org.
References
Author notes
Sequencing data of this study are available in the European Genome Phenome Archive (accession numbers EGAS00001005250 and EGAS00001001952).
Data are available on request from the corresponding author, Jun J. Yang (jun.yang@stjude.org).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal